Navigating Data Scarcity: AI's Emerging Role in Biotech
‘Garbage in, garbage out’ is a well-known principle in the machine learning (ML) community, and it is certainly true when it comes to adopting ML-based methods in biotech and drug discovery.
According to a recent McKinsey report, ‘lack of high-quality data sources and data integration’ was named as one of the three key factors slowing down digitalization and data analytics in life sciences (the other two being lack of cross-disciplinary talent, and lack of tech adoption at scale).
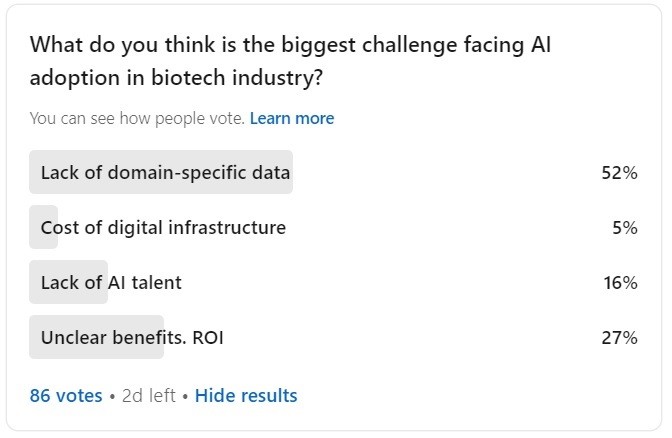
My own small poll here on LinkedIn resulted in the 52% of respondents favoring ‘lack of domain-specific data’ as the biggest challenge facing AI adoption in the biotech industry (a decent part of the respondents list are subject matter experts, based on my brief review).
Tackling the problem of data scarcity
San Francisco-based ‘techbio’ company Atomic AI developed a tool to tackle the lack of data about RNA structures.
Atomic AI’s proprietary AI-driven 3D RNA structure engine, known as PARSE, generates RNA structural datasets, integrating machine learning foundation models with large-scale, in-house experimental wet-lab biology to unveil functional binders to RNA targets.
The company’s technology has the ability to predict structured, ligandable RNA motifs at unprecedented speed and accuracy, a key barrier to current approaches to RNA drug discovery.
Atomic AI plans to use its database of discovered and designed 3D RNA structures to develop a pipeline of rationally designed small-molecule drug candidates.
What is interesting, Atomic AI is using so-called geometric deep learning, and can learn from very small RNA data.
Geometric deep learning is a subfield of machine learning that generalizes traditional neural network methodologies to data on non-Euclidean domains, such as graphs, manifolds, and complex networks. It seeks to understand data through its inherent geometric structures and relationships,
The method, called the Atomic Rotationally Equivariant Scorer (ARES), surpasses existing techniques in performance—even with training on just 18 known RNA structures. ARES's capacity to learn from minimal data addresses a significant challenge faced by typical deep neural networks. With its reliance solely on atomic coordinates and no RNA-specific details, this method has potential applications in various fields including structural biology, chemistry, and materials science, among others.
According to this Science paper, ARES operates without any predetermined ideas regarding the essential features of a structural model's accuracy. It doesn't come with any inherent understanding of double helices, base pairs, nucleotides, or hydrogen bonds. ARES's methodology isn't exclusive to RNA; it can be applied to any molecular system.
Instead of pre-defined specifications, the initial stages of the ARES network are tailored to detect structural patterns, learning their identities during training. Every layer calculates various characteristics for each atom, considering the spatial arrangement of adjacent atoms and the outcomes from the preceding layer. The only inputs for the initial layer are the 3D coordinates and the chemical element classification of every atom.
Zero-shot Learning
Another interesting example of tackling the data problem in biology was demonstrated by Canadian company Absci, focusing on designing antibodies using AI.
Absci has pioneered a milestone in generative AI for drug development by being the the first (as they claim) to craft and verify therapeutic antibodies using zero-shot machine learning.
What's zero-shot?
It's a machine learning approach where a model is trained on certain categories of data and is then able to make predictions or classifications on entirely new, unseen categories, often leveraging the relationships between known and unknown categories. For example, if trained on images of horses, the model might be able to recognize zebras, even if it hasn't been explicitly trained on zebra images.
In Absci’s case, antibodies are designed to latch onto certain targets without any prior training data from known antibodies for those targets.
Why is this significant? The zero-shot model by Absci produces antibody configurations distinct from existing antibody databases, encompassing de novo versions of all three heavy chain CDRs (HCDR123), the antibody regions most critical to target binding.
How efficient is this approach? In tests against over 100,000 antibodies, Absci’s success rate proved to be between five and 30 times higher than established biological benchmarks.
Synthetic data
A quite innovative concept is the application of synthetic data to close the data gaps in those areas where real data is scarce. What is synthetic data?
Synthetic data is information that's artificially manufactured rather than generated by real-world events, but it has probability distribution similar to the real data. It, therefore, can be used for training machine learning models the same way as real data.
For instance, there is promising evidence that state-of-the-art synthetic data models can produce artificial versions of even highly dimensional and complex genomic and phenotypic data.
Researchers from Gretel.ai, in collaboration with Illumina’s Emerging Solutions, are investigating the possibility of generating synthetic versions of real-world genomic datasets. The synthetic data crafted by Gretel preserves the structure of the original dataset while ensuring increased privacy, allowing researchers open access without jeopardizing patient confidentiality. Initial studies on a sample of 1,220 mice have shown promising results, suggesting that synthetic data can potentially revolutionize data sharing in genomics. Gretel and its collaborators aim to further refine the scalability, accuracy, and privacy of synthetic genomics data in the future.