A Brief Guide To Assay Technology For Efficient Drug Discovery -- Part 1
Effective drug discovery begins with the right assay, but the definition of "right" will shift as technology advances. More often than not, "right" is the product of tribal knowledge, namely the traditions of one's close peer group, study lineage and corporate culture. Instead, the right assay should be a fit-for-purpose application born of a broader, continuously updated, and unbiased consensus. As Steve Hamilton, aka The Lab Man, at the Society for Laboratory Automation and Screening (SLAS) has often stated in his blog posts, "developing assays – properly – is the cornerstone for life sciences R&D."
Those new to screening and lead discovery may struggle to know where to start, while veterans can always use a refresher course. In this review, which will appear in four installments, I will share my approach to staying current with trends in discovery assay technology. Assay ontology resources geared toward assay information management are the place to start (as Part I of this series).
By far the most broadly recognized compendium is the Assay Guidance Manual, a continuously updated eBook. It came to life as a Lilly effort to compile the tribal knowledge within its therapeutic project team silos, then rapidly evolved to provide guidelines for measurements of new and known molecular entities written by consensus among scientists in academic, non-profit, government and industrial research laboratories. NIH/NCATS has also been a motive force here. The full spectrum of applications to support SAR is covered, whether target or phenotype driven. These include biochemical, functional and, now more so, all the cell imaging modalities, including microscopy, that integrate into machine learning (AI) assisted discovery aids. The 2017 "what's new" list, for example, handily illustrates the scope and breadth of authorship in this indispensable publication.
For an encyclopedic but, less editorially guided view of assay diversity both the ChEMBL and PubChem's BioAssay databases are public repositories that cover 500,000 plus assay protocols searchable by concept, target, or molecular structure of expected drug-like structures. The features of PubChem BioAssay appear to dominate interest, as reviewed recently (O/A) in the SLAS Discovery journal. Both resources do a creditable job of breaking out assays into logical classification trees, although the PubChem BioAssay architecture offers a more succinct capture of the relevant information about assay types. This latter task is also accomplished in the SLAS LabAutopedia and, even more thoroughly, in the BioAssay Ontology (BAO) and BioAssay Research Database (BARD) portals.
BAO is an interesting tool for formalizing the domain of biological screening assays into a broader knowledge discovery framework connected to PubChem. It is an extensible, knowledge-based, highly expressive description of biological assays making use of descriptive logic based features of the Web Ontology Language (OWL). The BAO team has made their latest source code openly available. It has evolved considerably over the last six years since the original concept publication (O/A) by Visser and colleagues at the Department of Computer Science, U. Miami.
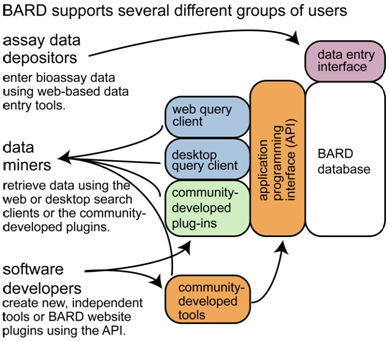
The better known of the two portals, and now under the aegis of NIH, BARD originated in a large academic consortium and report (O/A), lead by Clemons and collaborators in the Center for the Science of Therapeutics at the Broad Institute. NCATS and one of the key designers of BAO were also part of the operational team with the shared goal to specifically provide access and a set of tools to manipulate bioassay data from the NIH Molecular Libraries Program, with particular emphasis placed on the description of assay protocols. The database's current scope includes case materials on 631 projects, 4111 assay definitions, 6158 experiments and applications involving 221 chemical probes. The Java-centric BARD source code and instructions for building local implementations are shared openly as well via GitHub.
At this juncture two questions may be asked. Short of working through information overload on biochemical, functional and cellular assay diversity, does a one page, summary exist? If one takes a look at cancer drug discovery as a representative model, reasonably so because it is the largest field within discovery screening, then Hall's team at NCATS provides an answer to this first question in a recent study (O/A) titled "Small-Molecule Screens: A Gateway to Cancer Therapeutic Agents with Case Studies of Food and Drug Administration-Approved Drugs." The distribution of assay technologies historically applied to cancer-relevant assays, which appears to mirror the current universe of discovery assays in general, is shown pictorially in Figure 4. Of 295 annotated assays, 146 were biochemical, predominated by fluorescence and fluorescence polarization techniques, and 149 were cell-based, again with over-representation by luciferase reporter gene, luminescence and fluorescence readouts.
The companion question, for which there is no easy answer, calls out what types of more traditional assays have been under-represented, especially for orthogonal confirmation of hits without resorting to imaging or genomic techniques. Digging through all these resources looking at assay frequencies indicates that despite the wealth of current applied knowledge, "old-fashioned" but content rich biophysical measurements have been relatively neglected in the popular palette of bioassay tools. Fortunately, there is a way to play catch up, as pointed out in now multiple blog posts. This recondite aspect of the literature is succinctly reviewed by Folmer in the context of high throughput screening and by Renaud and colleagues taking a broader perspective in Nature Reviews Drug Discovery. The expectation is reasonable that information on this relevant methodology will soon start percolating through the compendial platforms discussed above.
In the next installments as separate posts, I will cover the more forward looking approaches in early stages of adoption, respectively, with regard to cell imaging, new generations of combined proteomic/transcriptomic single cell screening, and open access advances in assay data information management.
Topics: Emerging Technologies