Virtual Immunostaining for Digital Pathology
Owkin is a French-American startup, backed by Google Ventures among others, that deploys Artificial Intelligence (AI) and Federated Learning for medical research. The company was co-founded in 2016 by Thomas Clozel, a hematologist oncologist and researcher, and Gilles Wainrib, a computer science teacher-researcher at the École Normale Supérieure, and a Stanford University PostDoc.
Over the years, the team at Owkin has developed AI systems to analyze and interpret multimodal medical data, including pathology images, radiology images, genetic data, lab analysis, and clinical outcomes. The company recently announced the launch of a new product -- Virtual Staining, integrated as a feature in Owkin Studio platform.
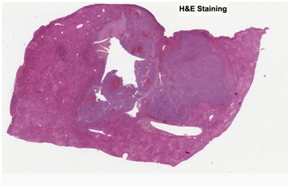
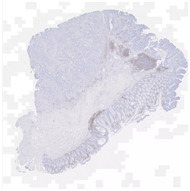
Hematoxylin and eosin (“H&E”) stain is the histology gold standard for medical diagnosis. H&E stained specimens show cell nuclei in blue and the extracellular matrix and cytoplasm in pink. To determine which cell types are present in a sample, pathologists will often combine H&E staining with immunohistochemistry stainings (IHCs) to selectively identify antigens. This enables pathologists to understand the distribution of specific cell types in the sample, providing crucial information to determine diagnosis and tumor grade.
|
|
|
|
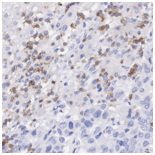
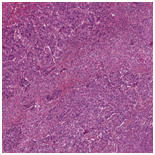
H&E stained whole slide image |
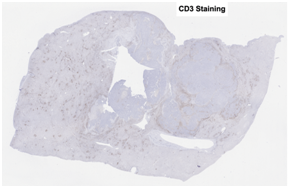
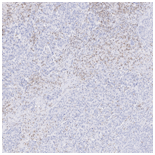
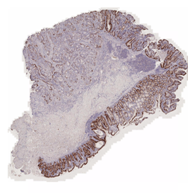
CD3 stained whole slide image (IHC) |
One common diagnostic biomarker used in IHCs is CD3, a co-receptor for T-cell activation. The CD3 immunostain can detect normal and neoplastic T cells as well as immune infiltration of the tumor microenvironment. This stain offers huge prognostic value, as T-cell activation is linked to overall immune response.
Obtaining an image of a CD3 immunostained sample requires specialist laboratory equipment and procedures, typically carried out by expert contract research organizations or dedicated service labs within hospitals. The output is an image that pathologists can analyze on a computer.
Owkin has developed a deep learning based data enrichment model that can produce a virtual stain of CD3 from existing H&E whole slide images, thereby removing the need for any laboratory procedures to generate IHC images.
To achieve this, Owkin adapted the supervised “image to image” translation pix2pix algorithm to work on whole-slide images. The model was trained on 50,000 pairs of aligned tiles (one H&E, one CD3) and validated on 50,000 pairs of tiles.
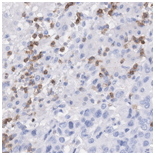
The virtual CD3 staining model was successfully able to automatically detect T cells from the generated CD3 images without any expert labeling. When comparing the average amount of CD3 immunostaining in tiles between the original "physical” and model generated validation tiles, we obtained a Spearman correlation of 93.6%. This indicates that virtual CD3 staining can be confidently used to quantify local variations in CD3 expression across the slide.
|
|
|
|
|
|
|
|
|
Virtually Stained CD3 image |
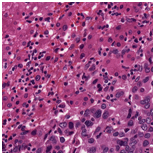
Original H&E stained image |
Original CD3 stained image |
Immunostainings can be a time consuming and costly process, particularly when applied on large sample banks. Furthermore, immunostainings are typically not available in retrospective studies or for existing datasets as the slides may not be available or have since been destroyed.
The virtual staining platform has the potential to positively impact researchers in both academia and industry in many different ways.
First, it provides a way to rapidly generate additional biomarker data for existing data banks for H&E slides. This provides prognostic value for translational researchers and pathologists by enriching existing data and improving the prediction of patient survival and response to treatment to aid patient treatment decisions.
Second, to enrich retrospective datasets with additional markers. This can significantly enhance the value of a dataset by identifying specific new biomarkers related to T cell density as potential therapeutic targets for example.
Finally, the virtual staining platform could also be used to enhance clinical trial design in a rapid and cost-effective manner, by using routine H&E histopathology to prospectively select high-value subgroups of patients with specific protein or immune profiles.
For more information, please visit https://owkin.com/virtual-staining/ where you’ll find a whitepaper and video demonstration of the technology.
|
|
|
|
|
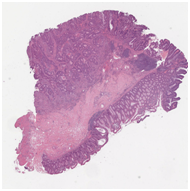
Unseen TCGA H&E slide |
Virtually stained CD3 |
Virtually stained AE1/AE3 |
In this article, we presented the results of the virtual staining platform as applied to CD3 in liver and colon tissue samples. The platform extends to other tissue types, as well as additional stainings including but not limited to AE1/AE3 for epithelial cells, CD163 for macrophages, CD15 for granulocytes, CD38 for plasmocytes, CD20 for B-cells, and CD117 for mastocytes.