Arc Institute Opens Virtual Cell Challenge With $175K in AI Benchmarking Prizes
The Arc Institute has launched the Virtual Cell Challenge, a new open competition to benchmark computational models that predict how human cells respond to genetic perturbations. Announced today and published in Cell (Vol. 188, Issue 13), the challenge invites AI and computational biology researchers to submit predictive models for scoring on a live leaderboard, using single-cell RNA sequencing data from human embryonic stem cells.
See also: Arc Institute Releases its First Virtual Cell Model
The inaugural edition centers on context generalization: participants must train their models using data from one set of cell types, then predict gene expression responses to CRISPRi perturbations in a held-out cell type—the H1 human embryonic stem cell line. The core dataset, newly generated by Arc for the competition, includes ~300,000 single-cell profiles spanning 300 CRISPRi perturbations, sequenced with high depth (~50,000 UMIs per cell) and cell coverage (~1,000 cells per perturbation) using the 10x Genomics Flex platform.
Participants will use perturbation-response data for 150 training genes to build their models. A validation set of 50 held-out perturbations allows real-time scoring on the public leaderboard, while a final test set of 100 additional perturbations will be released on October 27, with final submissions due November 3. Winners will be announced in December 2025.
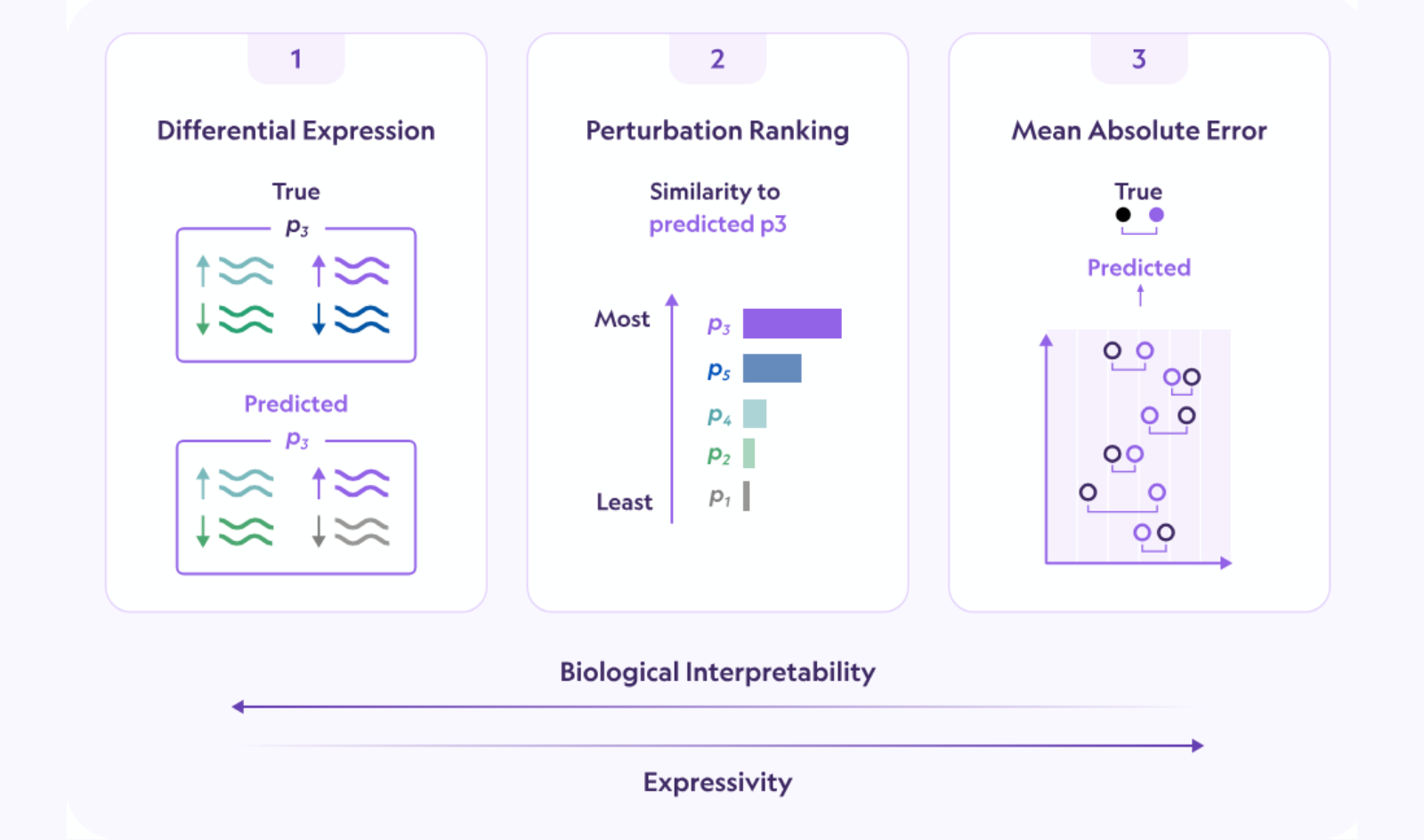
Model predictions are evaluated using a composite score combining three metrics:
- differential expression score (how accurately a model captures differential gene expression)
- perturbation discrimination score (how well a model distinguishes effects of different perturbations)
- mean absolute error (quantifying global expression profile accuracy).
All metrics are weighted relative to a baseline derived from the training set's cell-type averages.
Source: Arc Institute
Arc also provides additional public resources to aid model development, including its scBaseCount database (300M+ single-cell profiles across 72 tissues and 26 organisms), the Tahoe-100M dataset of ~100 million drug perturbation profiles, and multiple published perturb-seq datasets.
See also: Vevo Therapeutics Open-Sources Largest Single-Cell Dataset with Arc Institute
The top three performers will receive awards valued at $100,000, $50,000, and $25,000, respectively—each split between cash and cloud GPU compute credits. The competition is backed by NVIDIA, 10x Genomics, and Ultima Genomics, and will recur annually with increasing biological complexity and new datasets.
More information and registration: https://virtualcellchallenge.org
Topics: AI & Digital