Profluent Publishes AI-Designed Genome Editor in Nature, Releases Largest CRISPR Dataset
Profluent, an AI-first protein design company founded in 2022 in Berkeley, CA, has published research in Nature on OpenCRISPR-1, an AI-designed genome editor capable of editing the human genome. The study presents OpenCRISPR-1 as a viable alternative to SpCas9, showing reduced off-target effects, lower immunogenicity, and comparable or superior editing efficiency.
OpenCRISPR-1 was created using large language models trained on over 26 terabases of genome data. This effort enabled the discovery of 1.2 million CRISPR–Cas operons, forming the CRISPR–Cas Atlas, the largest known dataset of CRISPR systems. From this, Profluent used its models to generate 4 million novel CRISPR–Cas proteins, a 4.8× increase in diversity across Cas9, Cas12a, and Cas13 families.
See also: Profluent Unveils AI Model to Advance CRISPR-Cas Systems for Gene Editing
Among these, OpenCRISPR-1 stands out with 403 amino acid differences from SpCas9 and 182 differences from any known natural Cas9, underscoring its novelty. Structural predictions using AlphaFold2 showed that over 80% of generated proteins had high-confidence folds (pLDDT > 80).
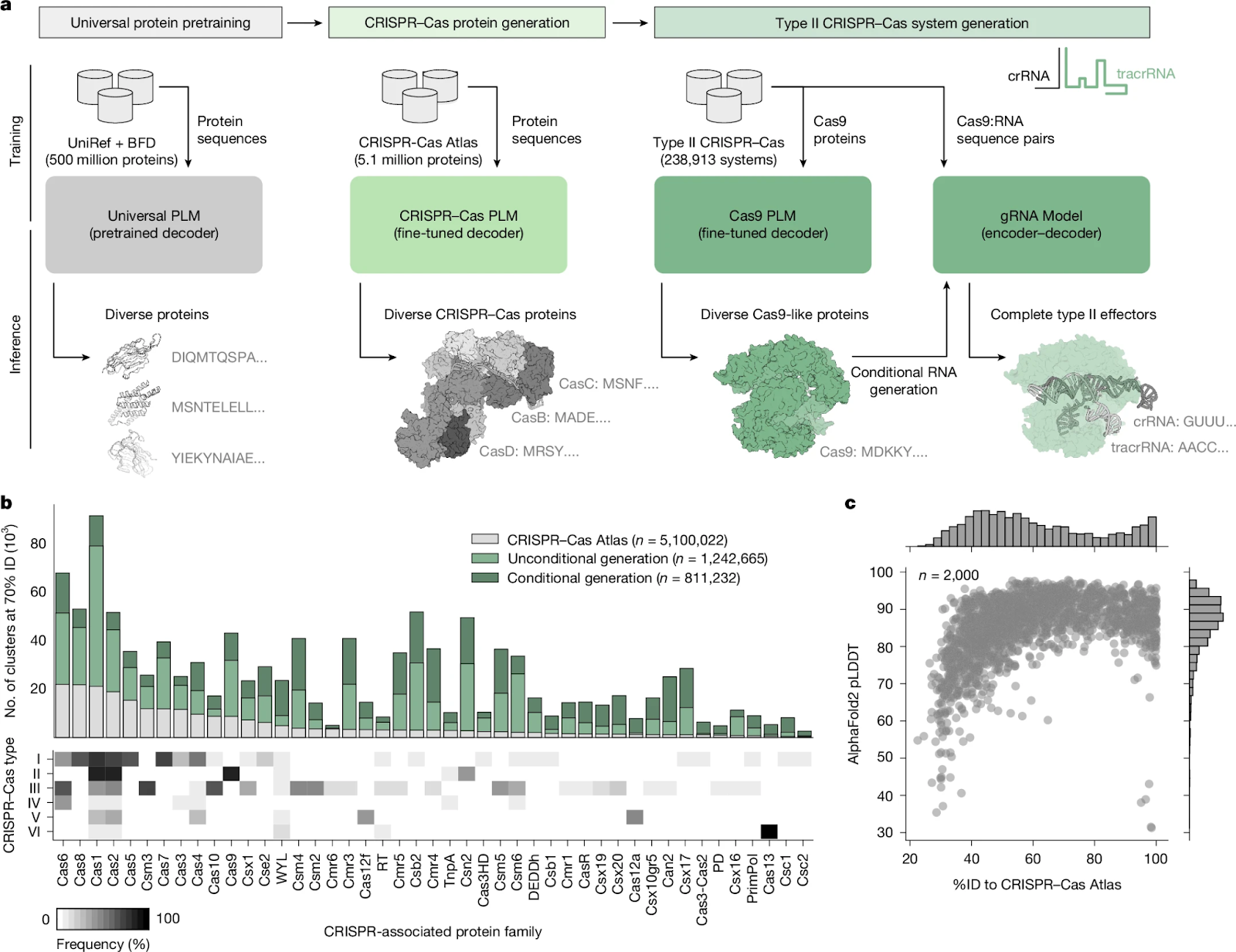
Generation of diverse Cas protein families; from Nature paper
Key performance findings:
- Genome-wide SITE-Seq assays showed significantly fewer off-target effects compared to SpCas9.
- iELISA tests using serum from multiple donors indicated significantly lower immune response for OpenCRISPR-1 than for SpCas9.
- OpenCRISPR-1 is compatible with base editing, enabling A-to-G conversions using both existing and AI-generated deaminases.
- A dedicated gRNA model co-designed tracrRNA and crRNA sequences specific to novel proteins, with custom sgRNAs improving editing efficiency at targeted loci.
- Unlike traditional structure-based design, Profluent’s language model approach produced structurally complete and functional proteins without overfitting.
Since its initial release in 2024, the OpenCRISPR-1 sequence and the CRISPR–Cas Atlas have been made publicly available for research and commercial use. The sequence is accessible via GitHub and AddGene. The full CRISPR–Cas Atlas dataset is now also available on GitHub, along with supporting code and models.
Code and pretrained parameters for the underlying protein language models were used according to instructions in their respective repositories: ProGen2 and ESM-2. Fine-tuned model checkpoints for the CRISPR–Cas and Cas9 LMs are available on Zenodo, and code for PAM and tracrRNA compatibility as well as gRNA design is also available on Zenodo.
Topics: AI & Digital