Chai Discovery Releases All-Atom Foundation Model for Zero-Shot Antibody Design with 16–20% Hit Rates
Chai Discovery has released Chai-2, a generative molecular design model for zero-shot antibody and binder discovery. The model is capable of designing fully de novo CDRs based solely on a given epitope and target structure, without reliance on existing antibody templates or training-time scaffolds. The company reports double-digit experimental hit rates (usually between 16–20%) validated across 52 previously unaddressed antigens using 20 candidates per target in a standard 24-well-plate format.
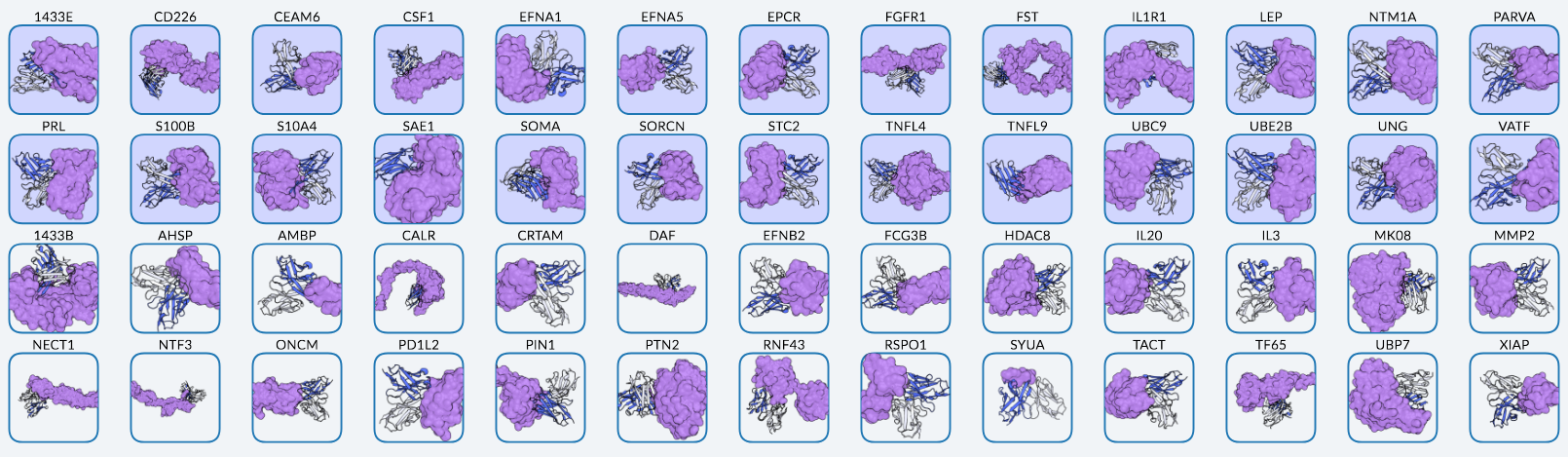
Chai Discovery: "52 novel antigens targeted by Chai-2. Blue boxes indicate targets with at least one successful binder out of ≤20 assayed designs, representing 50% of the tested targets."
Context
Chai was founded in early 2024 and released its first model, Chai-1, in September that year. At the time, Chai-1 demonstrated a 77% success rate on the PoseBusters benchmark, and did not rely on multiple sequence alignments (MSAs)—a common but computationally expensive technique used in many protein structure prediction models. Instead, Chai-1 achieved high-accuracy structure predictions from single sequences.
The company raised nearly $30 million at a $150 million valuation just six months after founding, with investors including OpenAI, Thrive Capital, and Dimension. Chai’s team includes veterans of OpenAI, Meta FAIR, Google X, and Stripe.
See also: 19 Companies Pioneering AI Foundation Models in Pharma and Biotech
New Model
Chai-2 combines all-atom structural modeling with a generative design model to produce antibody sequences. Instead of relying on large screening libraries or directed evolution, it generates new sequences that differ significantly from known antibodies and can be applied to difficult target sites. The system supports design across antibody modalities (scFv, VHH nanobodies) as well as miniprotein scaffolds. For miniproteins, Chai-2 achieved a 68% hit rate across five test targets, all confirmed in wet-lab validation.
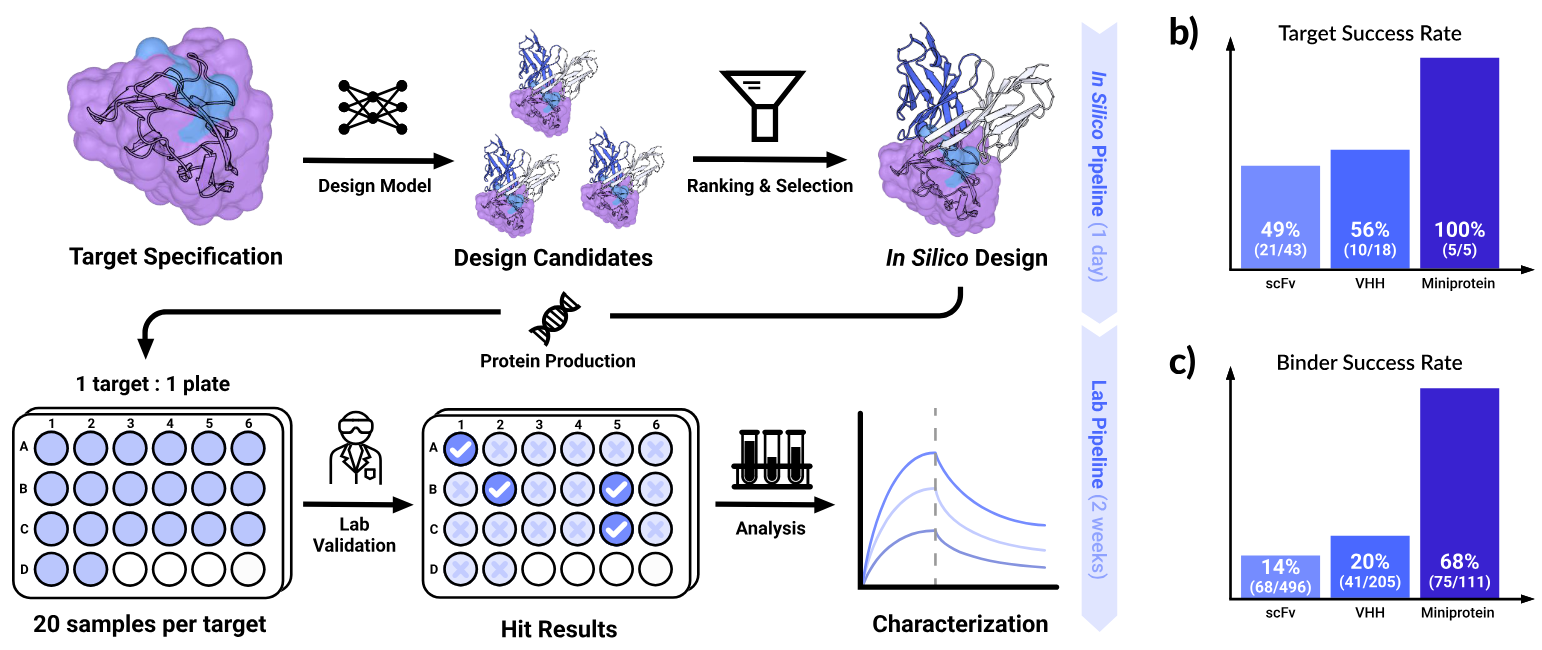
Chai-2 Technical Report, Figure 1: Chai-2 generates binder candidates from a target structure and epitope, ranks them in silico, and advances top designs to biochemical assays in a 24-well format. The full design-to-validation cycle takes about two weeks. Panel (b) shows the fraction of targets with at least one validated binder for each modality (scFv, VHH, miniprotein). Panel (c) shows the overall binding rate across all tested designs per modality.
Notably, the model generalizes beyond conventional biologics: it can reason over ligands and post-translational modifications, and is designed to extend to macrocycles, enzymes, and small molecules by modeling interaction potential at the atomic level. The platform also supports more complex design tasks like multi-specific and cross-reactive binders.
Collectively, we see our results as establishing computational-first design as an integral component of modern discovery platforms.—Chai Discovery Team in Chai-2 Technical Report
Limitations and Roadmap
While Chai-2 presents strong initial results in zero-shot antibody design, the technical report outlines several experimental constraints and future directions that frame both the model’s capabilities and its current boundaries:
- Format-specific results: all binding data in this study were collected using scFv and VHH formats. The authors note that affinities may differ when reformatted to Fab or full-length mAb constructs due to steric or conformational effects.
- Partial therapeutic profiling: although binding and specificity were characterized, the team has not yet completed full assessments of thermal stability, aggregation propensity, viscosity, or in vivo immunogenicity. These remain areas of active development.
- Hard-target validation: among the benchmarked targets is TNFα, which is considered computationally intractable due to its flat, polar interface. Chai-2 generated binders to it, providing a practical test of model generalization.
- Advances in structure prediction: Chai-2 doubled the rate of high-accuracy antibody–antigen complex predictions over its predecessor, Chai-1, achieving DockQ > 0.8 for 34% of test cases, underscoring improved atomic-level modeling.
- CDR loop complexity: despite these gains, modeling of antibody CDR loops remains a known bottleneck due to their flexible, diverse conformations—an acknowledged limit compared to the more rigid scaffolds of miniproteins.
- Toward zero-shot therapeutics: the team envisions future models that incorporate design constraints for manufacturability, pharmacokinetics, viscosity, and expression, aiming to computationally generate IND-ready biologics in a single in silico pass.
Vision
Chai's long-term goal is producing IND-ready biologics in a single in silico pass. The company describes this shift as a transition from stochastic to programmable discovery. To support safety and dual-use governance, Chai is granting access to academic and industry users selectively under its Responsible Deployment policy, which prioritizes programs with clear health and societal benefit while restricting work on unsafe or ethically concerning molecular designs.
See also: 17 More Biomedical Foundation Models
Topic: AI in Bio