How Latest Drug Candidates Were Discovered? (Retrospective Review, 2016-2017)
Recently D.G.Brown and J.Boström of AstraZeneca published an insightful analysis, where they reviewed lead generation research strategies behind 66 small molecule clinical candidates published over 2016-2017 in Journal of Medicinal Chemistry.
Below is a brief summary of some key statistics and ideas outlined in the work (I encourage reading the original paper, it contains a ton of valuable insights and a strong list of references).
Lead generation strategies
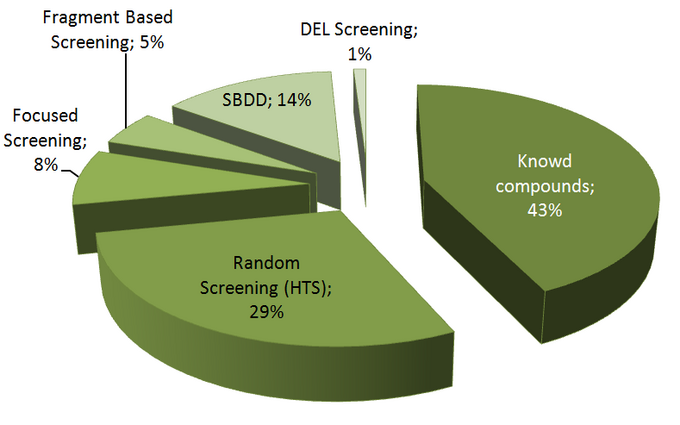
The authors found that 43% of drug candidates were ideated out of previously known starting points from literature, patent specifications, or previous programs.
Random screening (HTS) of large compounds collections (up to millions compounds) was second most efficacious category having produced 29% of clinical candidates, while focused screening and fragment-based lead generation approaches generated only 8 and 5% of drug candidates, respectively.
Knowing the 3D structure of a protein target is always a substantial advantage and so structure based drug design (SBDD) helped produce as much as 14% of clinical lead candidates (not taking into account general in silico efforts used in most other hit-generation strategies).
DNA-encoded library screening (DEL) approach, which included screening of the largest chemical libraries up to a billion of molecules with DNA-tags attached to them, is largely falling behind other research strategies with only 1% of clinical lead candidates produced in this period (which is quite in accordance with the fact that DEL is available to a very limited number of pharma and CROs, and also there are some technical issues in the way to be yet solved.)
Interestingly, the overwhelming majority of all clinical candidates in this study was found to be discovered using target based approaches, while only a couple of compounds originated from phenotypic screens without any prior knowledge about a particular target class -- and they primarily were identified in the infection-based screens.
Targets and therapeutic areas
Probably not surprisingly, around 30% of all clinical candidates came from kinases as targets, followed by GPCR’s (17%), epigenetic targets (9%, an emerging class of targets), and other enzymes, including phosphodiesterases, transferases, proteases, etc.
Other target classes included ion channels (9%), nuclear receptors and protein-protein interactions (both around 1.5%), and miscellaneous categories (9%).
Authors found that most clinical lead candidates were in the areas of oncology (30%), CNS/Pain (18%), infection diseases (13%) and metabolic diseases (12%), while such therapeutic areas as Immune/inflammatory, cardiovascular, respiratory, urological and hematological diseases were all falling behind.
How close were hits and clinical candidates?
Authors analyzed hits originating from two biggest R&D categories -- from known compounds and random screening -- and found out that physicochemical properties and structural characteristics changed in a different manner.
For example, both categories demonstrated an increase in MW by 84 and the number of heavy atoms by 6 on average, and an increase in the number of rotatable bonds (+1.2 on average), while no significant change in the clogP was observed when going from the initial hit molecule to a clinical candidate molecule.
In most cases, the optimized compounds were structurally very different from the hit compounds -- both in the case of using random HTS and previously known compounds as a guiding light. Typically, lead optimization stage would result in adding four aliphatic and two aromatic atoms, and two hydrogen-bond acceptors (HBAs), with a general increase in the polar surface. In contrast, the numbers of hydrogen bond donors (HBDs) and fraction-sp3 did not change significantly.
Below is a random example showing how drastically a hit molecule can be altered during the lead optimization stage:
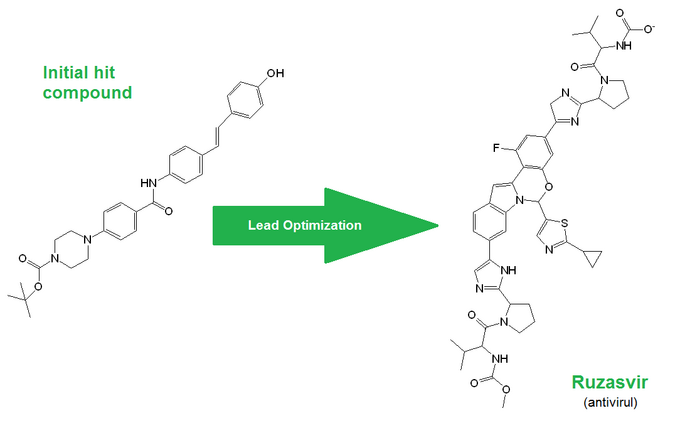
Strikingly, in 66% of all cases, at least one heterocyclic nitrogen atom was added during the hit-to-lead optimization stage, and in 30% of cases at least one fluorine atom was added during the development stage.