Isomorphic Labs Presents an AI Drug Design Engine That Goes Beyond AlphaFold 3
On the heels of Johnson & Johnson multi-modality drug discovery collaboration last month, Isomorphic Labs has now published benchmark results for its unified AI drug design engine, the Isomorphic Labs Drug Design Engine (IsoDDE), positioning it as a unified, multi-model system that extends beyond AlphaFold 3 into drug-design tasks including generalisation-heavy protein–ligand structure prediction, antibody–antigen modelling, binding affinity prediction, and ligand-blind pocket detection.
According to an Isomorphic Labs spokesperson, where AlphaFold focuses on the question of what a protein looks like, IsoDDE moves toward answering how to design a drug that can bind to it and whether that drug is likely to have a therapeutic effect.
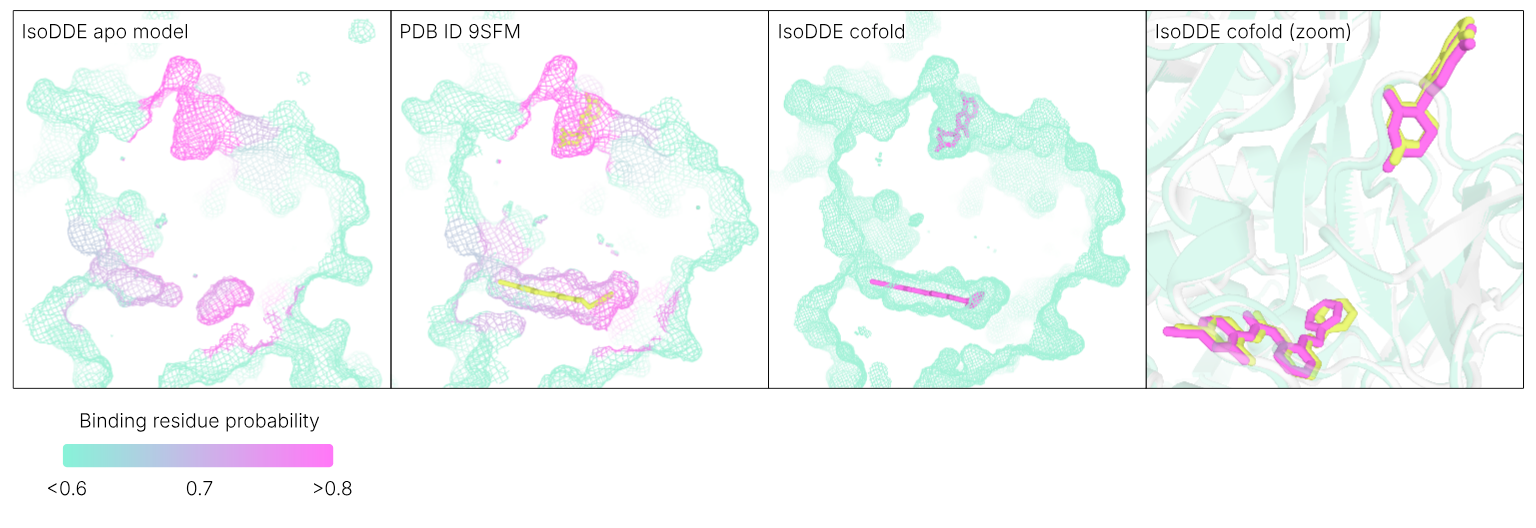
IsoDDE was able to recapitulate the recent discovery of a novel cryptic site on cereblon (Dippon et al., 2026) predicting the location of both the known and the novel cryptic sites using just the sequence of cereblon as input, without specifying the identity of the ligands. Credit: Isomorphic Labs
A subset of IsoDDE’s capabilities is previewed below and detailed further in the accompanying technical report.
AlphaFold 3, built by Isomorphic Labs together with Google DeepMind, advanced the field by predicting 3D structures of proteins, including DNA, RNA and small molecules, giving scientists a clearer view of how biological components fit together, but its strength was mainly structural.
Benchmarks later showed that AlphaFold 3’s accuracy dropped for structures unlike those in its training set, meaning it performed best on cases resembling what it had already seen. This highlights the difficulty of generalizing into uncharted areas of biomolecular space, where one finds many of drug discovery’s toughest problems and key opportunities.
See also: 14 Startups in AI Protein Design: Platforms, Specialists, Modular Tools
As reported by Isomorphic Labs, IsoDDE shows improved generalization to protein-ligand structures far from its training set. On the 'Runs N’ Poses' benchmark, designed to test out-of-distribution performance, (Škrinjar et al. 2025) it more than doubles AlphaFold 3’s accuracy on the hardest cases. The model successfully captures complex phenomena like induced fit and cryptic pocket opening, even in systems highly dissimilar to those seen during training.
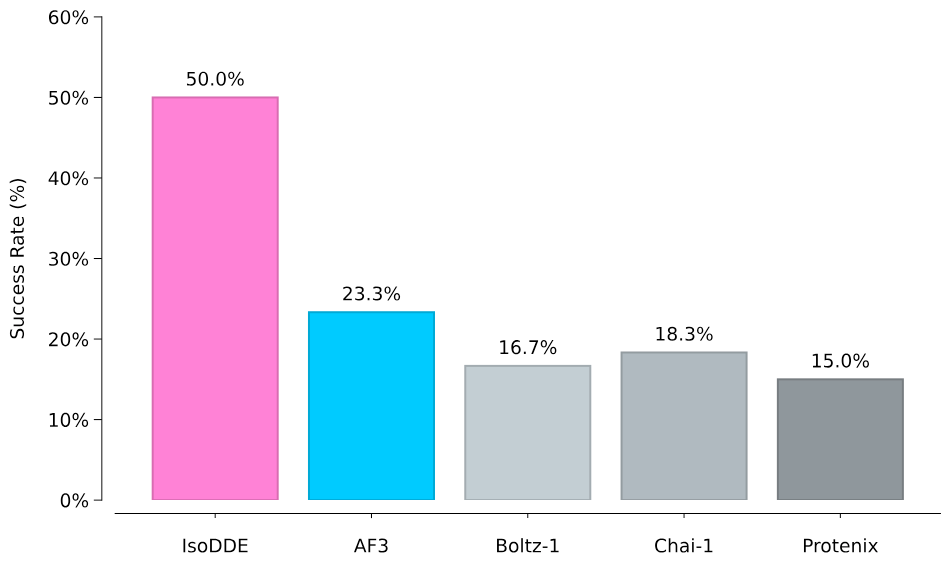
Protein-ligand structure prediction accuracy on hardest generalisation category ((0-20])- most dissimilar to the training set - from the 'Runs N’ Poses' benchmark. Credit: Isomorphic Labs
IsoDDE also focuses on antibodies, which have large, flexible, loop-shaped binding surfaces. One of the hardest regions for any model to predict is a CDR-H3 loop, which plays a central role in how an antibody recognizes its target and varies a lot in length and sequence, making its 3D shape and how it sits on the antigen especially difficult to model accurately.
On a set of 334 low-similarity antibody–antigen complexes, IsoDDE consistently generated interface structures more similar to the real, experimentally observed ones than both AlphaFold 3 and Boltz-2, using comparable computational resources. It also demonstrated strong performance specifically in modeling the CDR-H3 region.
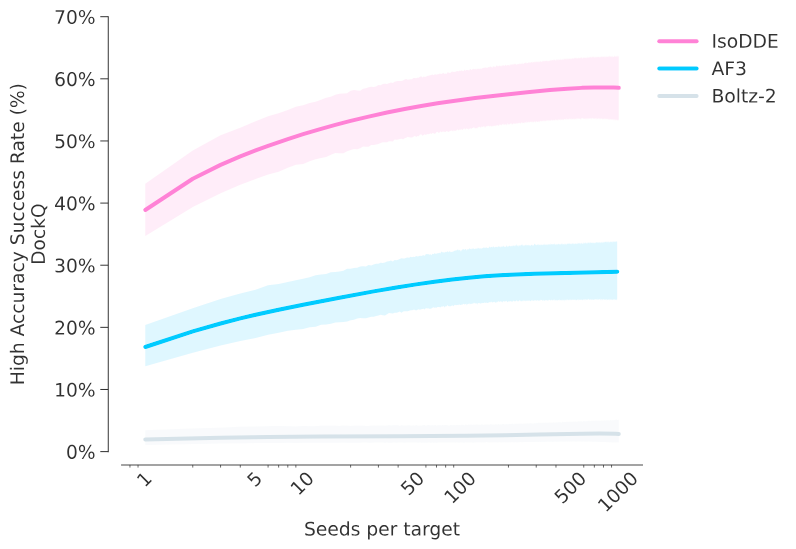
Antibody-antigen structure prediction accuracy for a challenging, low-homology test set (𝑛=334), showing the fraction of high-quality predictions at the protein interface (DockQ >0.8) as inference-time compute is increased. IsoDDE outperforms AlphaFold 3 by 2.3x and Boltz-2 by 19.8x. Credit: Isomorphic Labs
Traditional free-energy perturbation methods like FEP model atomistic motions to estimate binding affinities but are compute-intensive and depend on high-quality experimental structures. Notably, IsoDDE is reported to outperform other deep-learning models on three public affinity benchmarks and, in some settings, to match or exceed FEP accuracy without requiring crystallographic starting structures.
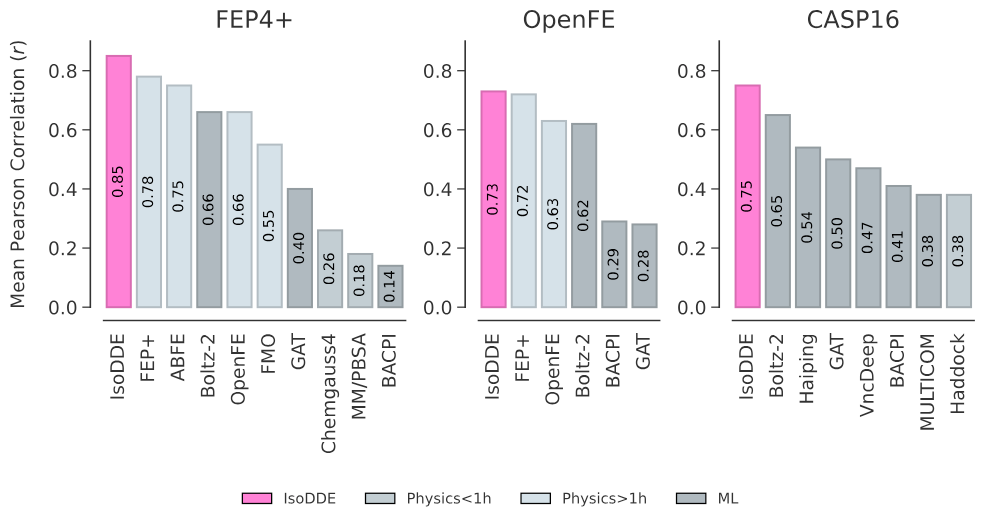
Binding affinity prediction performance across a range of public benchmarks. Credit: Isomorphic Labs
According to the Isomorphic Labs’ post, the engine is already being used by in-house drug discovery teams to:
- Explore “unseen” protein structures for which little experimental data exist.
- Search for and prioritize binding pockets on new or poorly characterized targets.
- Generate and rank new small-molecule and biologic candidates based on predicted structure and affinity.
London-based Isomorphic Labs, founded by Alphabet Inc. in 2021 to apply DeepMind's AI breakthroughs to drug discovery, has established itself as a fast-rising actor in computational drug discovery. With high-profile partnerships, including multi-target deals with Novartis, Eli Lilly, and now Johnson & Johnson, Isomorphic is positioning AI as a central engine in modern drug design, bridging machine learning and pharmaceutical R&D at scale.
In July 2025, then-President Colin Murdoch said Isomorphic’s internal pipeline included oncology and immunology candidates, and that first-in-human clinical trials were “very close.” This year, at the World Economic Forum in Davos, CEO Demis Hassabis said Isomorphic Labs now expects its first AI-designed drugs to enter clinical trials by the end of 2026, pushing back the company’s earlier goal of starting trials in 2025.
Topic: AI in Bio