Terray Launches Experiment-Driven Machine Learning Platform for Small-Molecule Discovery
Terray Therapeutics introduced EMMI—Experimentation Meets Machine Learning—as an integrated platform that pairs ultra-dense microarray experimentation with a full AI stack used daily across its discovery programs. The company reports more than 13 billion proprietary compound-target measurements generated through its microarray system, expanding by roughly 1 billion per quarter.
According to the latest company’s insight, these data support internal immunology programs and partnerships with Calico, Bristol Myers Squibb, Gilead, and Odyssey Therapeutics.
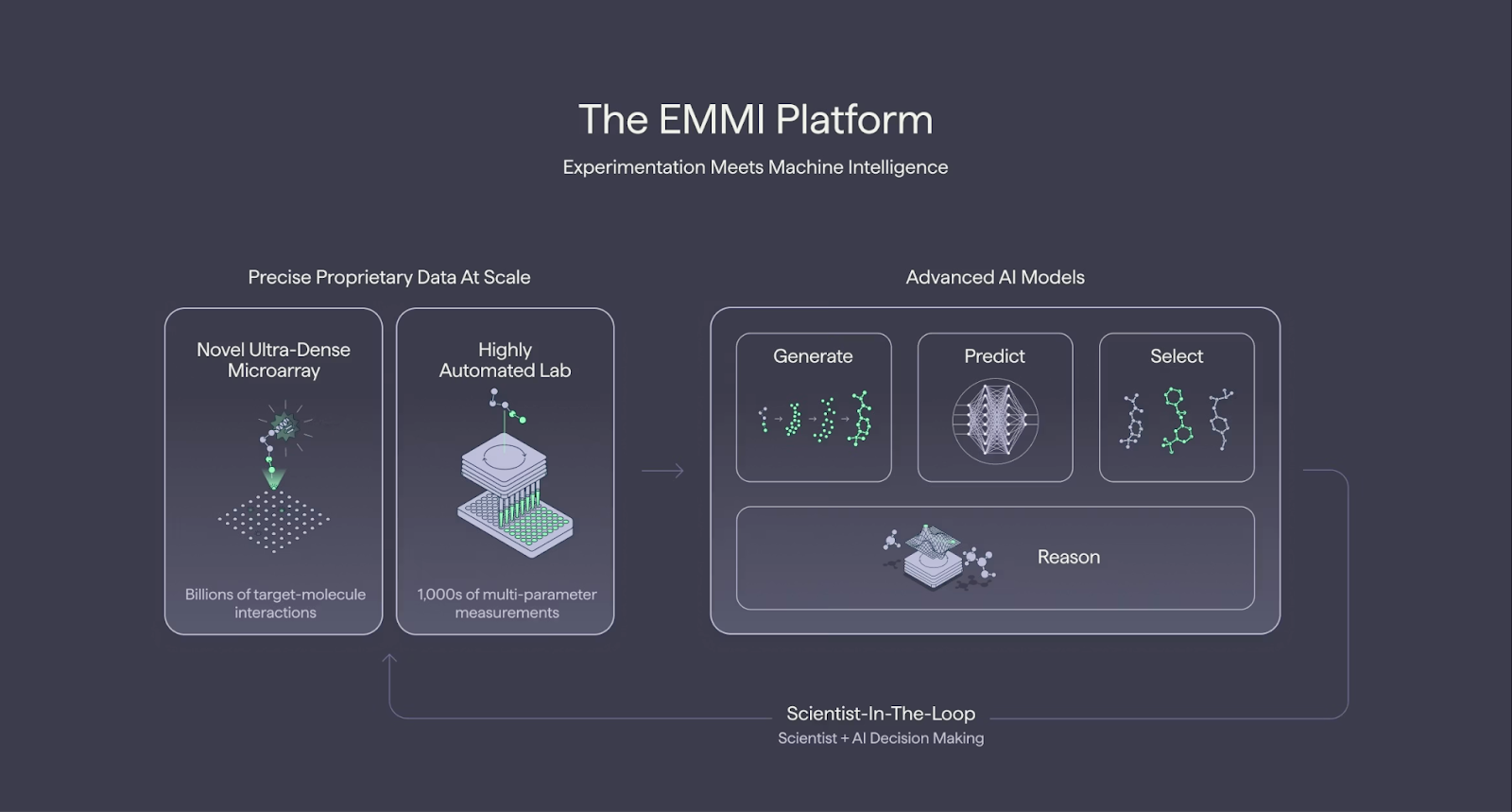
Image credit: Terray Therapeutics
COATI: Foundation Model for Molecular Understanding
At the center of Terray’s AI platform is COATI, a large-scale molecular foundation model that gives algorithms a way to understand and design molecules in the same way language models process text. Trained on more than one billion molecular structures, COATI links multiple chemical representations like SMILES strings, 2D graphs, and 3D conformers through an invertible mathematical mapping that allows the system to move seamlessly between understanding and generating molecules with specific properties.
Using contrastive learning, COATI learns relationships among different molecular formats, enabling inverse design where new molecules can be created to match desired biological or chemical traits. The latest version, COATI3, provides a 768-dimensional molecular representation. Training at this scale is supported by NVIDIA’s DGX Cloud, which supplies the GPU infrastructure required for such compute-intensive tasks, and the model’s framework is openly available on Terray’s GitHub repository, as well as a supporting JCIM paper.
Generative and Predictive Models
Terray’s generative models create new virtual molecules designed to have specific chemical or biological properties. They use two main AI methods—latent diffusion and reinforcement learning—to explore chemical space and propose molecules that balance potency, stability, and synthetic accessibility. The diffusion method focuses on fine-tuning properties using learned guidance, while the reinforcement learning approach directly optimizes for lab-ready molecules that can be made and tested more easily. In 2024, Terray introduced its first latent diffusion–based generative model built on COATI, using classifier guidance to fine-tune molecular properties, with source code available on GitHub.
Once thousands of potential molecules are generated, EMMI’s predictive models estimate which ones are most likely to succeed. Using Terray’s TerraBind potency model and data from its 13-billion-measurement database, these models predict how well molecules will bind to biological targets and how they might behave in the body. The system runs a two-step evaluation—first with an ultra-fast screening model, and then with a detailed structure-based model to identify the best candidates. Additional prediction tools assess key drug properties such as solubility, permeability, metabolism, and clearance, helping narrow the list to the few compounds most promising for synthesis and testing.
EMMI Select
The main technical announcement involves EMMI’s selection. The new EMMI Select module pairs Epistemic Neural Networks (Epinets) with an Expected Maximum (EMAX) acquisition function to quantify predictive uncertainty and guide experimental design, aiming to triple efficiency in potency optimization cycles compared with conventional approaches.
In drug discovery, molecule selection is a costly bottleneck, as chemists can synthesize only a limited number of candidates per week. Traditional strategies rely on score-based or diversity-based heuristics but ignore model uncertainty, often resulting in redundant or suboptimal compound batches. Terray’s approach uses Epinets to quantify confidence in molecular property predictions from its large-scale potency model, TerraBind, without requiring multiple model ensembles.
The associated preprint describing this implementation is available on arXiv, along with an accompanying open-source GitHub repository.
Broader Integration
Terray also plans to extend EMMI Select beyond potency optimization to multi-objective decision-making, incorporating ADME properties, synthesis cost, and assay time. This could support more integrated optimization across Design–Make–Test–Analyze (DMTA) cycles, potentially shortening preclinical iteration times and improving the selection process for development candidates. According to the company, the system is already being applied across internal and partnered pipelines to identify structurally novel candidates.
Topic: AI in Bio