DeepMind Introduces AlphaGenome for Predicting Effects of Human DNA Variants
A new AI model from Google DeepMind, called AlphaGenome, has been released in preview for non-commercial research, offering a high-resolution, long-context DNA sequence model designed to predict the regulatory effects of genetic variants at scale.
AlphaGenome is an artificial intelligence model trained to interpret how single-nucleotide variants (SNVs) and mutations in human DNA affect gene regulation. It builds on DeepMind's previous model, Enformer, and complements AlphaMissense, which focuses on protein-coding regions. AlphaGenome expands coverage to the 98% of non-coding DNA often implicated in regulatory processes and disease.
How It Works
The model takes DNA sequences up to 1 million base pairs long and predicts thousands of molecular properties that govern gene activity. These include transcription start and end sites, RNA splicing junctions, RNA output levels, DNA accessibility, and protein-DNA binding patterns, across diverse tissues and cell types.
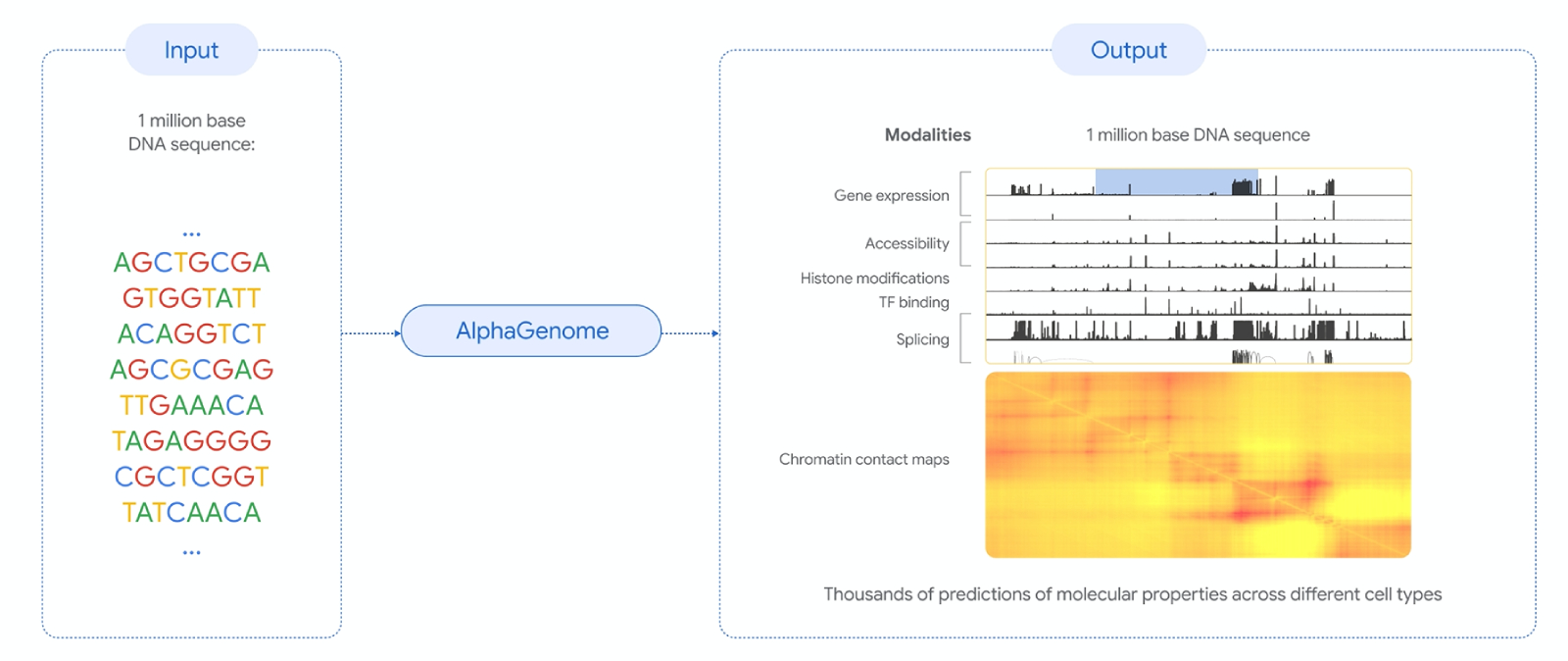
From the DeepMind blog animation: AlphaGenome processes a 1-million-letter DNA sequence to predict diverse molecular properties across tissues and cell types.
To do this, AlphaGenome combines:
- Convolutional layers to detect short sequence motifs,
- Transformer layers to process long-range dependencies across the entire DNA sequence,
- Task-specific heads to generate predictions for multiple modalities.
Training was conducted using publicly available experimental datasets from ENCODE, GTEx, 4D Nucleome, and FANTOM5 consortia, which profile gene regulation across hundreds of human and mouse cell types and tissues. A single model (without knowledge distillation) was trained in four hours using half the compute of Enformer, distributed across multiple Tensor Processing Units (TPUs).
Performance
AlphaGenome reportedly outperforms existing models across 22 of 24 single-sequence benchmarks and 24 of 26 human variant-effect prediction benchmarks. It is the first model to jointly predict all assessed regulatory modalities, including:
- Variant impacts on RNA splicing (relevant to diseases like SMA and cystic fibrosis),
- Long-range enhancer-promoter interactions,
- Effects on chromatin state and gene expression levels.
Unlike prior models, it delivers both long-range sequence context and single-base resolution without major trade-offs. It also allows efficient variant scoring by comparing mutated and unmutated sequences in under a second.
Potential applications include:
- Disease research: pinpointing regulatory variants involved in Mendelian or complex diseases,
- Synthetic biology: designing regulatory DNA for cell-type-specific gene expression,
- Functional genomics: mapping and characterizing the regulatory architecture of the genome.
In a test case, AlphaGenome successfully recapitulated the known mechanism behind T-ALL (T-cell acute lymphoblastic leukemia) by predicting that a non-coding mutation could activate the TAL1 gene via creation of a MYB motif.
AlphaGenome is not designed for personal genome prediction. It does not model disease traits involving higher-order biological factors, and performance drops for very long-range regulatory interactions (>100,000 bp). Future directions include expanding the model’s capacity for tissue-specific predictions and training on additional species.
Availability
AlphaGenome is available via API for non-commercial research use. Full release is planned later, with researchers invited to adapt and fine-tune the model for domain-specific tasks. The model is not validated for clinical use.
Topic: AI in Bio