Y Combinator Backs Yale Spinout Modeling Human Drug Response with AI To Reduce Preclinical Failure
CellType, a Yale University spinout, has joined Y Combinator’s W26 batch with a platform that combines foundation models of human biology with AI agents designed to run end-to-end AI modeling across discovery and early clinical strategy. The company positions its approach as a response to high preclinical failure rates, arguing that more faithful simulation of human biology could reduce attrition before clinical trials.
See also: Companies Applying AI to De-Risk Clinical Trials: 2026 Watchlist
The founding team includes David van Dijk, a Yale professor with publications in Cell, Nature, and ICML, and Ivan Vrkic, who co-developed the core modeling approach at Yale and previously led foundation model training at another biotechnology startup, and built software to control CERN's Large Hadron Collider.
CellType describes itself as an “agentic drug company,” integrating large-scale biological foundation models with task-specific AI agents that autonomously execute components of the discovery pipeline. The central premise is that many drug failures stem from preclinical systems that do not adequately reflect human biology. By simulating patient-level biological responses in silico, the company aims to evaluate therapeutic hypotheses before wet-lab screening and before first-in-human studies.
At the core of the platform is Cell2Sentence, a 27-billion-parameter foundation model developed at Yale in collaboration with Google DeepMind researchers. The model is trained on large-scale single-cell data and treats gene expression patterns as a form of biological language, converting cellular states into structured representations that can be analyzed computationally. The underlying idea is that, as with large language models, increasing scale allows the system to capture deeper structure in biology and potentially exhibit new capabilities beyond incremental performance gains.
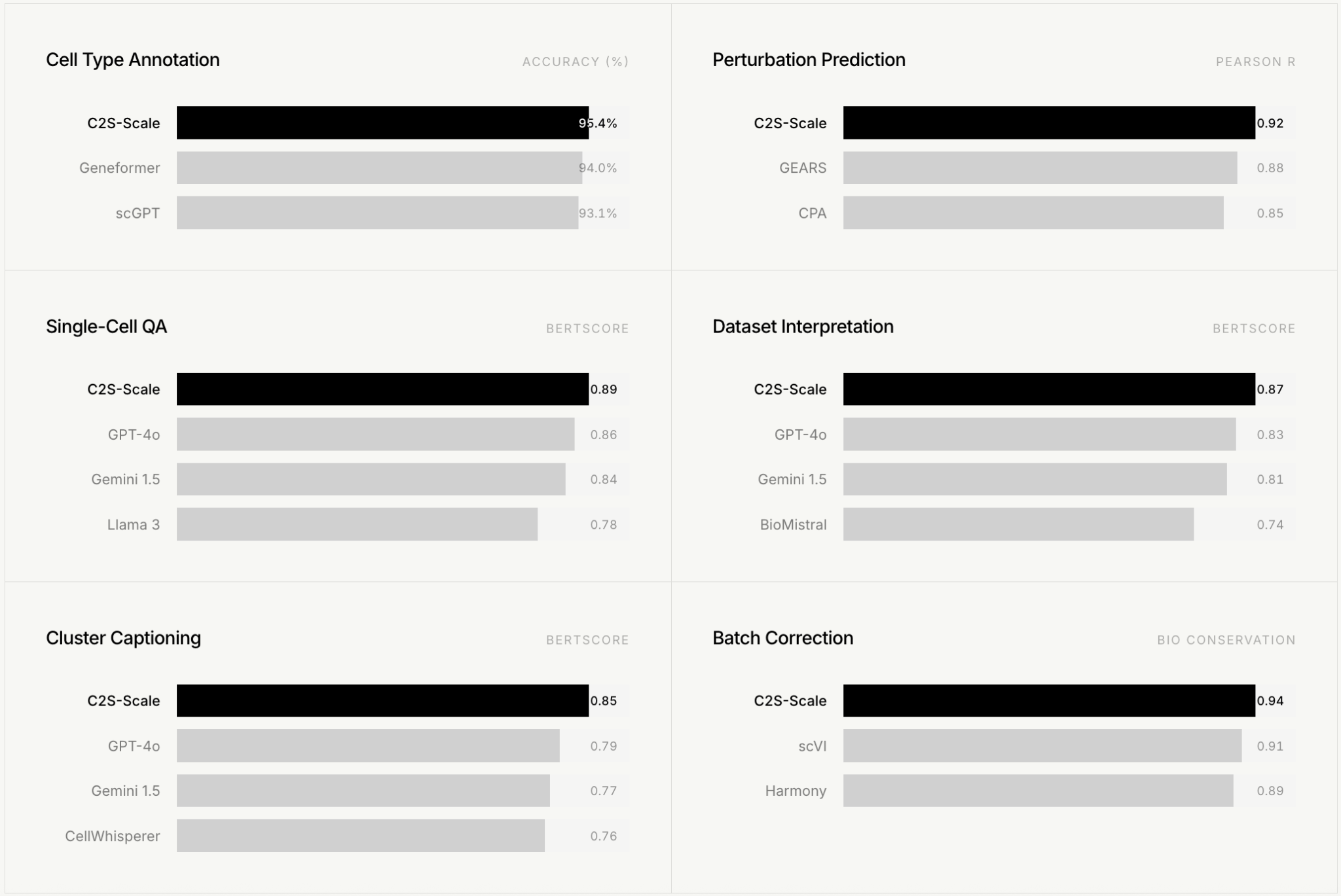
Results from C2S-Scale (2026). Evaluated on standard single-cell benchmarks. Image credit: CellType
To test this, the team applied the model to a problem in cancer immunotherapy: identifying drugs that could increase antigen presentation in tumors, but only under specific immune conditions. They constructed a dual-context virtual screen using patient-derived tumor samples with low interferon signaling and, separately, isolated cancer cell lines without immune context.
After simulating the effects of more than 4,000 drugs, the model prioritized compounds predicted to selectively enhance immune signaling only in the patient-relevant setting. A portion of the top-ranked hits had prior literature support, while others were previously unreported; the team reports that at least one cancer-related prediction was subsequently identified & validated in experiments in living cells.
Topic: Biotech Ventures