Terray Updates its Binding-Affinity Predictor with Diffusion-Free AI Architecture
Terray Therapeutics, a Los Angeles–based biotechnology company developing an AI-driven small-molecule discovery platform, has introduced a new version of its TerraBind potency prediction model within the EMMI platform. The company reports higher binding affinity prediction accuracy while substantially reducing computational cost.
Terray positions the update as addressing a core scaling bottleneck in AI-driven small-molecule discovery, where millions of candidate molecules must be evaluated efficiently during iterative design cycles.
The announcement builds on Terray’s previously disclosed EMMI stack (Experimentation Meets Machine Intelligence), which integrates high-throughput microarray experimentation with generative, predictive, and selection models trained on what the company reports as more than 14 billion compound–target measurements.
TerraBind functions as the central structure-based potency predictor in that workflow, supporting Design–Make–Test–Analyze loops across internal programs and collaborations with Bristol Myers Squibb, Gilead, Calico, and Odyssey Therapeutics. With Bristol Myers Squibb, the platform has already helped yield an AI-enabled discovery milestone.
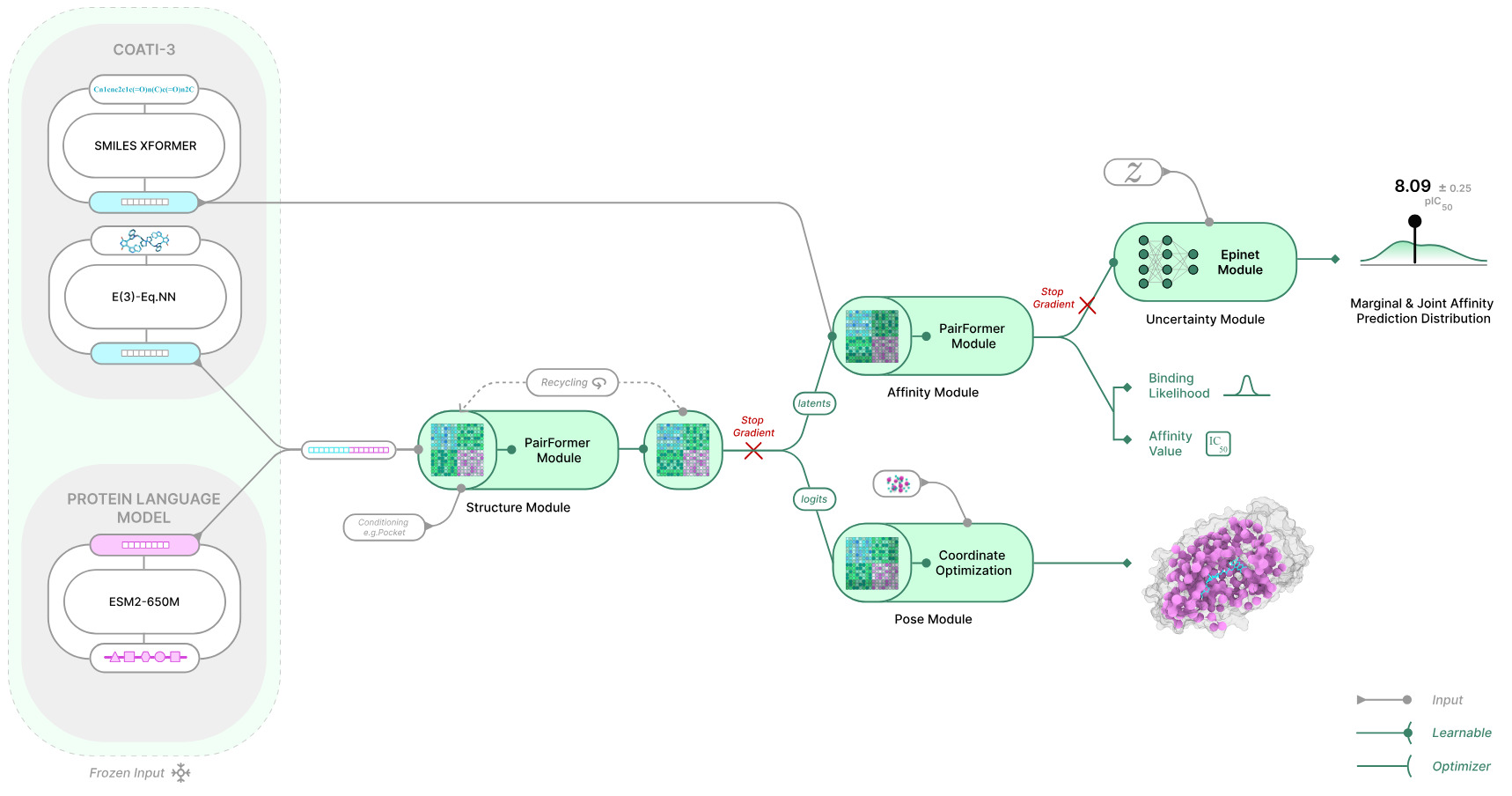
Figure 3: TerraBind Architecture. A modular, diffusion-free architecture combining pretrained protein and ligand encoders with pairformer-based structural and affinity prediction modules, including built-in uncertainty estimation. Image credit: TerraBind: Fast and Accurate Binding Affinity Prediction through Coarse Structural Representations, Matteo Rossi, Ryan Pederson et al.
Architecture Shift: Removing Diffusion from Binding Prediction
Terray says the speed and accuracy gains come from redesigning how the model predicts whether a drug molecule will bind tightly to a target protein.
Many recent AI models (like AlphaFold3-derived approaches and Boltz-2) first try to reconstruct a very detailed, atom-by-atom 3D picture of how a molecule fits into a protein. They use a technique called diffusion, which gradually refines a noisy structure into a precise one. This can produce highly detailed models, but it requires heavy computation and slows down screening when millions of candidate molecules must be evaluated.
TerraBind takes a different approach. Instead of modeling every atom, it simplifies the structure. It represents proteins by the central points of their amino acids and molecules by their key heavy atoms. Rather than generating a fully detailed 3D structure first, the model goes directly from this simplified representation to a binding strength prediction.
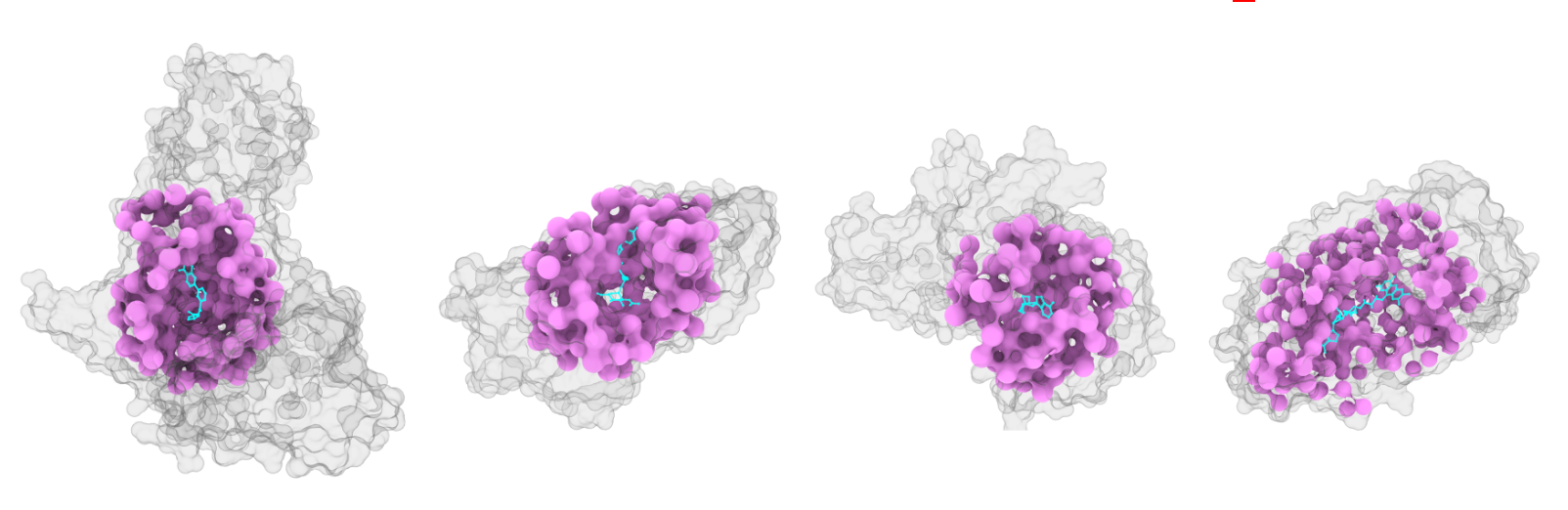
Figure 1: TerraBind coarse structure prediction. Image credit: TerraBind: Fast and Accurate Binding Affinity Prediction through Coarse Structural Representations, Matteo Rossi, Ryan Pederson et al.
The idea is that for estimating how tightly a molecule binds, full atomic detail may not always be necessary. By skipping the computationally expensive diffusion step, the model reportedly runs much faster and at much lower cost. Terray reports that this redesign makes TerraBind 26 times faster (according to structure-focused benchmarks such as FoldBench, PoseBusters, and Runs N’ Poses) and reduces inference costs by 96 percent compared with diffusion-based models, while maintaining competitive structural accuracy.
In side-by-side evaluations against Boltz-2, described as a leading publicly available comparator, Terray reports that TerraBind achieved a 16 percent higher correlation with experimental binding data on the CASP16 benchmark and an average 20 percent improvement across 18 internal drug discovery targets spanning multiple protein families and chemical scaffolds.
Topic: AI in Bio