Stanford–Arc Team Reports AI-Made Viruses That Kill Bacteria
Researchers at Stanford University and the Arc Institute report they used a genome-scale generative model to design novel variants of bacteriophage phiX174; 16 of 302 AI-proposed genomes replicated in E. coli and lysed bacteria, indicating functional “AI-written” viruses suitable for experimental use (MIT Technology Review, Sept. 17, 2025). Potential applications include phage therapy tools and viral vectors, alongside clear biosafety concerns around misuse.
See also: Evo 2: Largest Foundation Model for Genomic Research Across All Domains of Life
The work sits on top of Arc/Stanford’s “Evo/Evo 2” genomic language models, which were trained on large, multi-domain DNA corpora (Evo 1 was pretrained on ~2.7 million prokaryotic and phage genomes; Evo 2 expanded to ~9.3 trillion nucleotides across ~128k organisms) and are described as capable of sequence prediction and generative design at genome scale. Evo 2’s preprint details unconstrained generation up to chromosome and mitochondrial-genome lengths and open-sourced code/weights, contextualizing how a model could propose complete phage genomes rather than local edits.
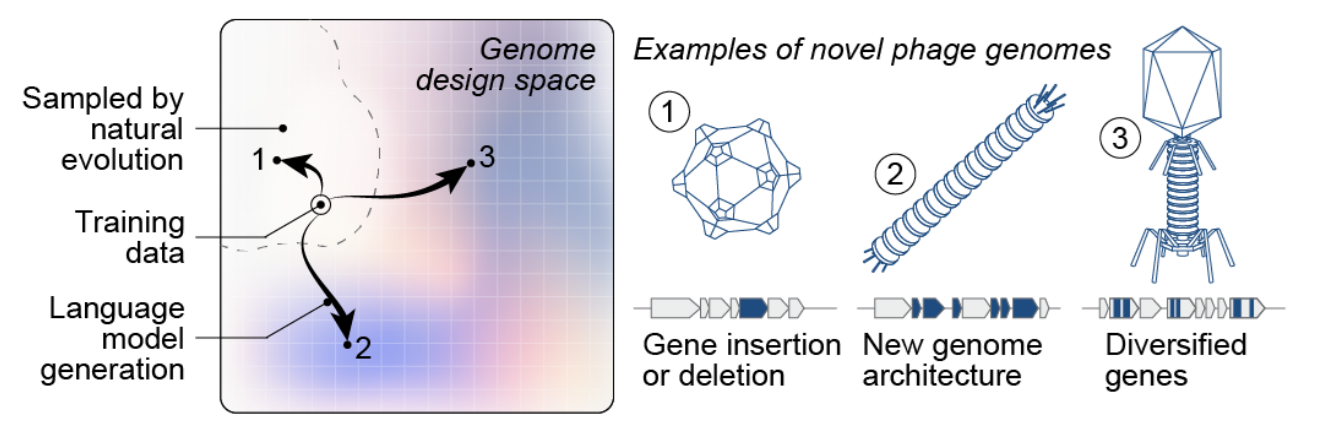
Generative genome language models have the potential to access novel phage genome design
In the new bacteriophage result, the team fine-tuned Evo models and generated ~300 φX174 genome designs; ~16 proved viable after synthesis and plating in E. coli. Sixteen designs formed plaques—i.e., the phage replicated and burst host cells—after which electron microscopy confirmed viral particles. While phiX174 is a small, well-studied ~5 kb, 11-gene ssDNA phage used historically in genome engineering, achieving replication with de novo genomic designs is a notable threshold toward “generative” genome engineering rather than mutation-level tweaks. (Methodologic and organism background: phiX174 genome features and use as a model; Evo/Evo2 capabilities.)
Safety and scope: the group says their training/data excluded human-infecting viruses; nevertheless, external observers flag that similar methods could be redirected to human pathogens, and that random enhancements pose risk. This mirrors broader debate around foundation models for genome design highlighted earlier this year when Evo 2 was introduced and discussed as enabling whole-genome sequence generation.
What’s new
-
Generative design at whole-genome granularity for a replicating virus. Prior phage-focused LLMs and Evo/Evo2 papers demonstrated long-context generation and simulated genomes; this report adds wet-lab replication and host lysis from AI-proposed, printed genomes.
- Throughput of proposals vs. functional yield. A 16/302 hit rate suggests non-trivial viability in whole-genome design space for small phages, with room to iterate using closed-loop lab automation. (Hit-rate and workflow per MIT Technology Review reporting.)
Why it matters (near-term)
Phage therapy and engineered delivery vectors are plausible near-term beneficiaries: phiX174-scale genomes are amenable to synthesis/assembly and rapid phenotypic screening, and generative proposals could expand viable sequence diversity beyond natural isolates. Evo 2’s public release and tooling lower barriers for academic labs to explore similar designs, increasing both scientific value and governance questions. This shift (from generative design to real-world utility) is something we're tracking closely on Where Tech Meets Bio.
Caveats: The bacteriophage study is a preprint-stage report according to MIT Technology Review; peer-reviewed validation and independent replication will be important. Scaling from 5 kb phage to bacterial or eukaryotic genomes remains a major step; even Evo 2’s demonstrations of longer sequences don’t remove assembly, compatibility, and “boot-up” constraints for cells.
Topic: AI in Bio