Potential Breakthrough in Cell Reprogramming as OpenAI and Retro Biosciences Report 50x Pluripotency Marker Expression Gains
OpenAI and Retro Biosciences have released new findings from a joint effort in AI-driven protein engineering. Using GPT-4b micro, a biology-specialized variant of GPT-4o, the team designed two modified versions of Yamanaka factors, RetroSOX and RetroKLF, that showed over 50-fold increases in pluripotency marker expression and improved DNA repair activity during cell reprogramming.
Yamanaka factors are a set of four proteins (Oct4, Sox2, Klf4, and c-Myc) that can turn back the “clock” on a cell’s identity. Introduced into adult cells like skin or blood cells, these factors reprogram them into induced pluripotent stem cells (iPSCs), cells that, like those in early development, can grow into nearly any tissue in the body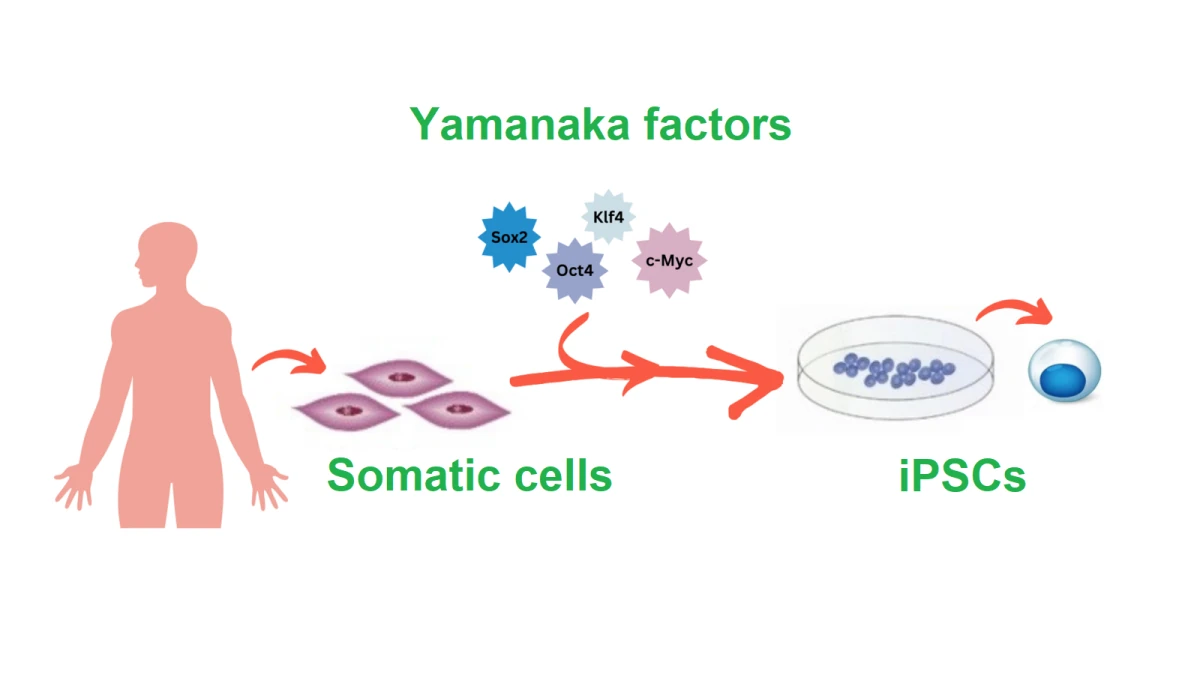
This process, first demonstrated by Shinya Yamanaka and collaborators in 2006, opened the door to regenerative therapies that don’t rely on embryonic stem cells. For aging researchers, the implications are significant: by reprogramming old or damaged cells, it may be possible to reverse cellular aging or repair age-related damage.
But the original Yamanaka cocktail comes with trade-offs, most notably, low efficiency and potential safety risks, especially from c-Myc, a gene linked to cancer.
The promise of Gen AI in Cell Reprogramming
GPT-4b micro was trained on protein sequences, biological texts, and tokenized 3D structural data to generate redesigned variants of Sox2 and Klf4.
The redesigned proteins, RetroSOX and RetroKLF, outperformed their wild-type counterparts across a range of biological tests.
Over 30% of AI-generated RetroSOX variants showed higher reprogramming activity than native SOX2, with some differing by more than 100 amino acids, while nearly 50% of RetroKLF variants exceeded manually engineered versions.
In fibroblast cells, these proteins led to faster and stronger expression of both early and late pluripotency markers, with indicators like NANOG and TRA-1-60 appearing days ahead of typical reprogramming timelines. Alkaline phosphatase staining confirmed more robust iPSC colony formation.
The approach was also tested in mesenchymal stromal cells from older adult donors, where over 85% of cells activated endogenous pluripotency genes within 12 days. Cells reprogrammed with the AI-designed proteins demonstrated full differentiation potential and maintained normal chromosomal structure. Notably, they also showed reduced DNA damage signals under stress, suggesting improved repair capacity during reprogramming.
In January 2025, OpenAI and Retro announced their collaboration and the development of GPT-4b micro for targeted biological applications. Retro Biosciences is reportedly raising a $1 billion in funding, including investment from OpenAI CEO Sam Altman.
The current results document the design and validation of modified transcription factors using a specialized AI model. Further studies are in progress to evaluate their suitability for preclinical and clinical research.
Topic: AI in Bio