LatchBio Publishes 25-Million-Cell Open-Source Spatial Transcriptomics Atlas
LatchBio has released a human spatial transcriptomics atlas of 25 million cells. Covering 45 tissue types, 63 diseases, and data from 11 major platforms (10X Genomics, Bruker CosMx, STOmics Stereo-seq, Vizgen MERSCOPE, Takara Bio Seeker, AtlasXOmics DBiT-seq, Spatial Genomics seqFISH, Element Biosciences AVITI24, and Singular Genomics G4X), the atlas is available via public data portal as annotated H5AD files. The developers generated datasets using “latch-curate”, a human-in-the-loop curation system that improves annotation quality by incorporating structured and unstructured study data.
See also: Building the Virtual Cell: AI Foundation Models and Billion-Cell Datasets
In parallel with atlas, LatchBio introduced a spatial toolkit for downstream analysis. Researchers can move from raw data through counts construction, image alignment, QC, differential expression, cell type annotation, and ligand-receptor analysis.
Partnerships with Takara Bio and AtlasXOmics provide streamlined pipelines, while visualization with Latch Plots supports interactive exploration of morphology and staining images. Advanced methods like cell2location and Squidpy are integrated, with region-of-interest tools for focused spatial interaction analysis.
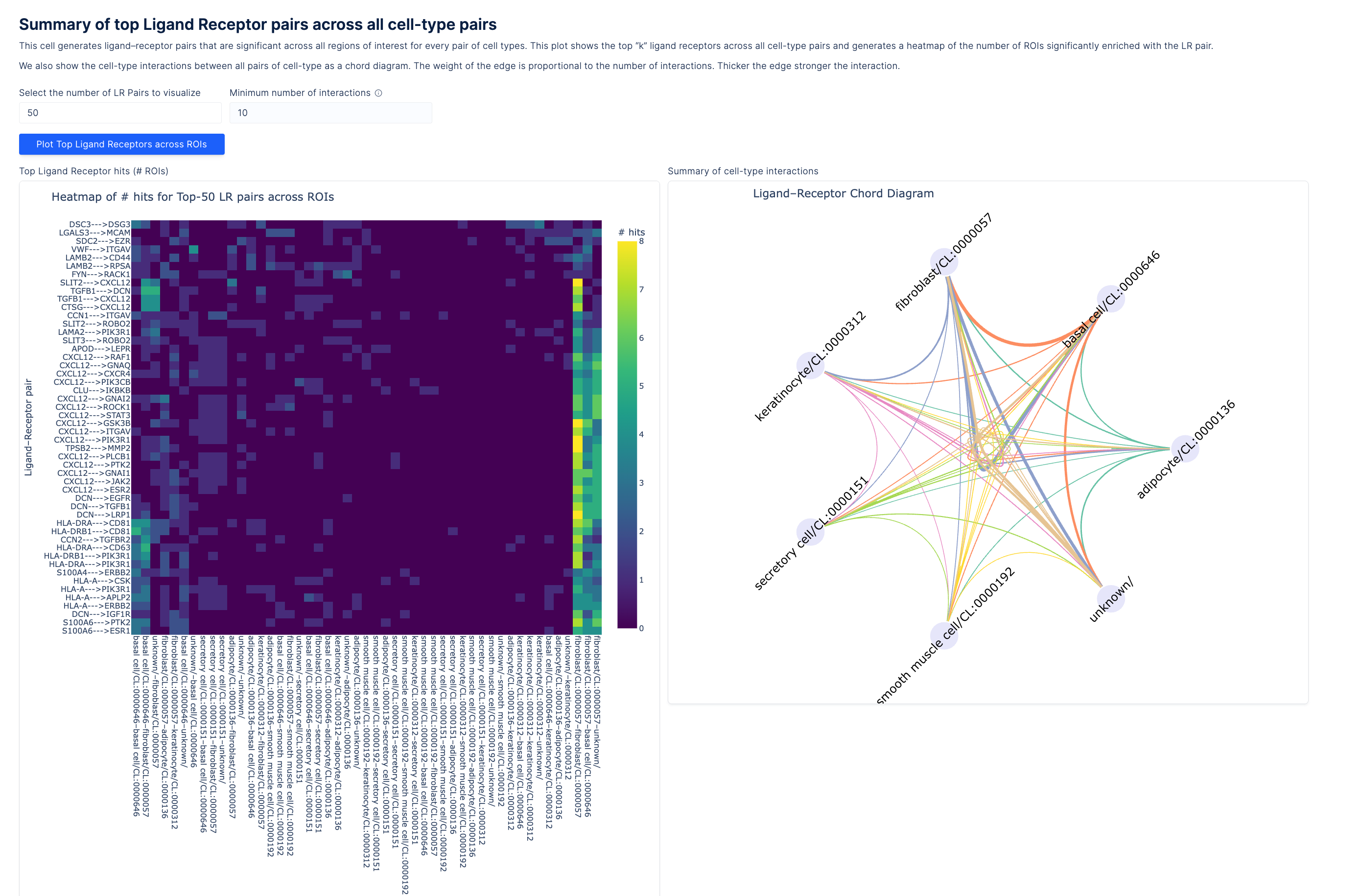
Ligand receptor pairs summary applying Squidpy
LatchBio, launched in 2021, targets providing the code-free data infrastructure for Biopharma R&D. The team, led by three former UC Berkeley students Alfredo Andere, Kyle Giffin and Kenny Workman, states the release of their cell atlas addresses lack of spatial organization, calling it the core bottleneck of previous single-cell focused large-scale initiatives.
Access the Dataset: https://console.latch.bio/data-portal