Immunai Offers Free Single-Cell Multi-Omics To Three Academic Centers While Growing Its AI Atlas
Immunai has selected researchers from Mount Sinai, Weill Cornell Medicine, and Massachusetts General Hospital to join its academic collaboration initiative focused on advancing understanding of the human immune system. The projects will generate data across key disease areas spanning autoimmune kidney disorders, solid tumors, and cell therapies.
Each team will receive access to Immunai’s sequencing and data analysis capabilities at no cost. This includes single-cell RNA sequencing and other multi-omic profiling, aimed at building deeper insights into how the immune system behaves in different conditions. Immunai plans to integrate these datasets into its AMICA platform, which supports its AI engine for immune system modeling and drug development.
AMICA (Annotated Multiomic Immune Cell Atlas) is Immunai’s central data resource, built from high-resolution single-cell sequencing of hundreds of thousands of patient samples. It combines proprietary and public datasets across more than 800 cell types and 500 diseases, using harmonized metadata and expert curation to support consistent analysis. AMICA integrates RNA expression, surface proteins, TCR/BCR repertoires, and spatial transcriptomics, forming a detailed immune reference for comparing patient data across conditions like cancer, autoimmunity, and infection.
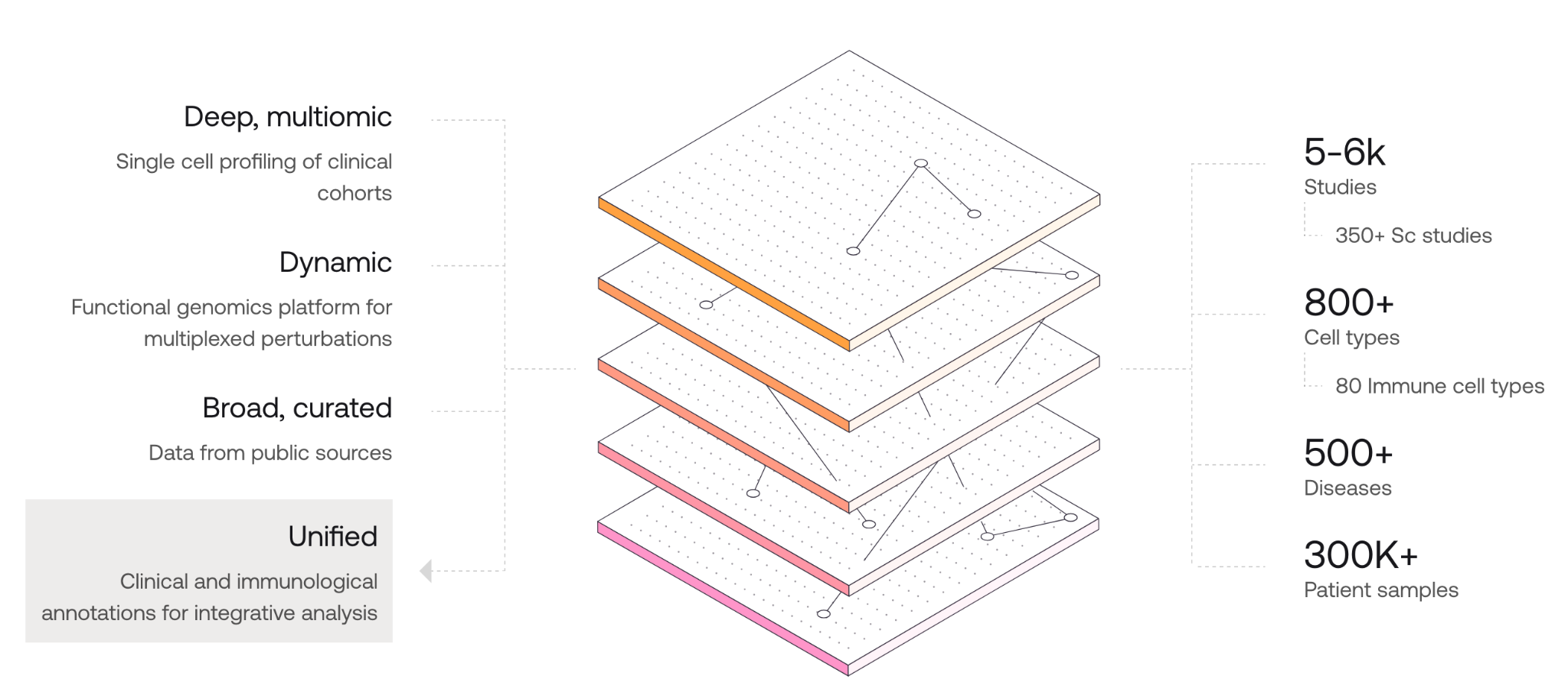
Image credit: Immunai AMICA
IDE (ImmunoDynamics Engine) is the AI layer that analyzes immune features within the AMICA dataset to extract mechanistic insights and clinical signals. It links immune cell behavior to treatment response, identifies biomarkers for patient stratification, and supports therapeutic hypothesis generation. IDE includes tools for modeling immune activation, validating findings in lab systems, and delivering clear recommendations through explainable output formats—helping researchers and clinicians make data-supported decisions in drug development and clinical trials.
The selected research projects include:
- Immune signatures in autoimmune kidney disease (Mount Sinai)
- Immune response tracking in T-cell therapy for multiple myeloma (Mount Sinai)
- Data from patients receiving neoadjuvant immunotherapy for lung cancer (Weill Cornell)
- Patterns of response and resistance in gastric cancer immunotherapy (Massachusetts General Hospital/Harvard)
The initiative is part of Immunai’s broader strategy to collect diverse, longitudinal immune datasets across diseases. After appointing former Pfizer R&D chief Mikael Dolsten to its board to guide oncology and immune discovery efforts in early 2025, in collaboration with the Parker Institute for Cancer Immunotherapy, Immunai started building what it describes as the largest single-cell dataset for real-world immunotherapy, covering 1,070 patients and 3,700 blood samples treated with checkpoint inhibitors. This multi-omic dataset feeds into Immunai’s AMICA atlas and supports biomarker and trial design via its IDE platform.
In August, the company opened its Grand Collaboration Initiative, offering free multi-omic sequencing for up to 1,000 academic samples. These efforts followed Immunai’s multi-year partnerships with AstraZeneca and Teva to apply its platform in oncology, immunology, and trial optimization.
Topic: Next-Gen Tools