New AI Reportedly Surpasses AlphaFold 3 in Drug-Protein Binding Prediction
Genesis Molecular AI, in collaboration with NVIDIA, has announced a generative 3D foundation model for protein–ligand structure prediction, Pearl (Placing Every Atom in the Right Location). Genesis reports state-of-the-art results on public benchmarks and internal datasets.
The company says performance gains are driven by large-scale physics-generated synthetic data, a new SO(3)-equivariant diffusion architecture, and NVIDIA’s cuEquivariance kernel accelerations; the model is already being deployed across internal and partner programs.
The announcement also formalizes a rebrand: Genesis Therapeutics is now Genesis Molecular AI. The a16z-backed company says it has raised over $300 million to date and is deploying Pearl at industrial scale across its Bay Area headquarters and other sites.
Genesis positions Pearl as a practical tool for everyday drug design rather than a demo model. In tests against AlphaFold 3 and open-source cofolding replicas (Boltz-1, Boltz-2, Chai-1, Protenix) using standardized checks for both accuracy and basic physical validity, Pearl reportedly led across all benchmarks—about 15% relative improvement over AlphaFold3 on Runs N’ Poses and up to 40% on PoseBusters.
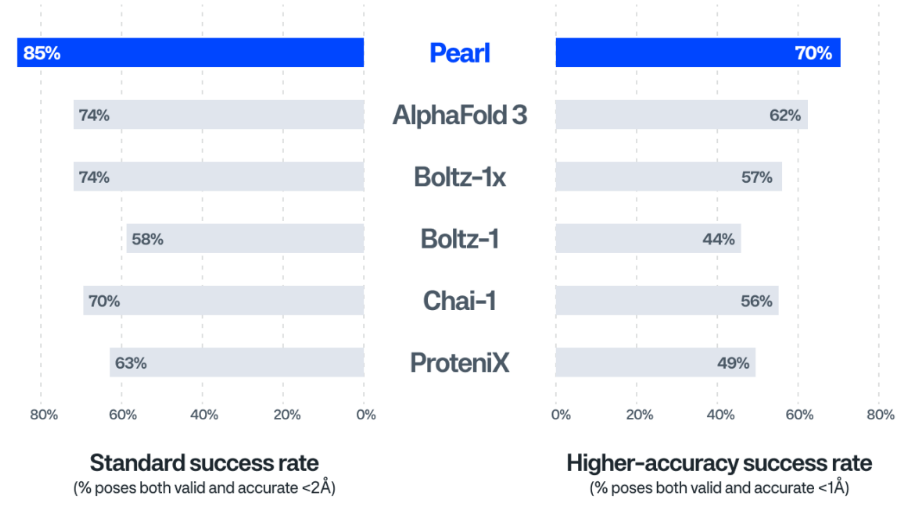
Genesis: Runs N' Poses benchmark, cofolding
The approach centers on two ideas: add a large volume of physics-generated synthetic protein–ligand structures to counter scarce experimental data, and pair that with a geometry-aware diffusion architecture that treats rotations as equivalent to the same 3D object. Genesis reports clear “scaling law” behavior—the model keeps improving as more simulated complexes are added—framing synthetic data as a lever the field can keep turning without waiting for slow, costly crystallography.
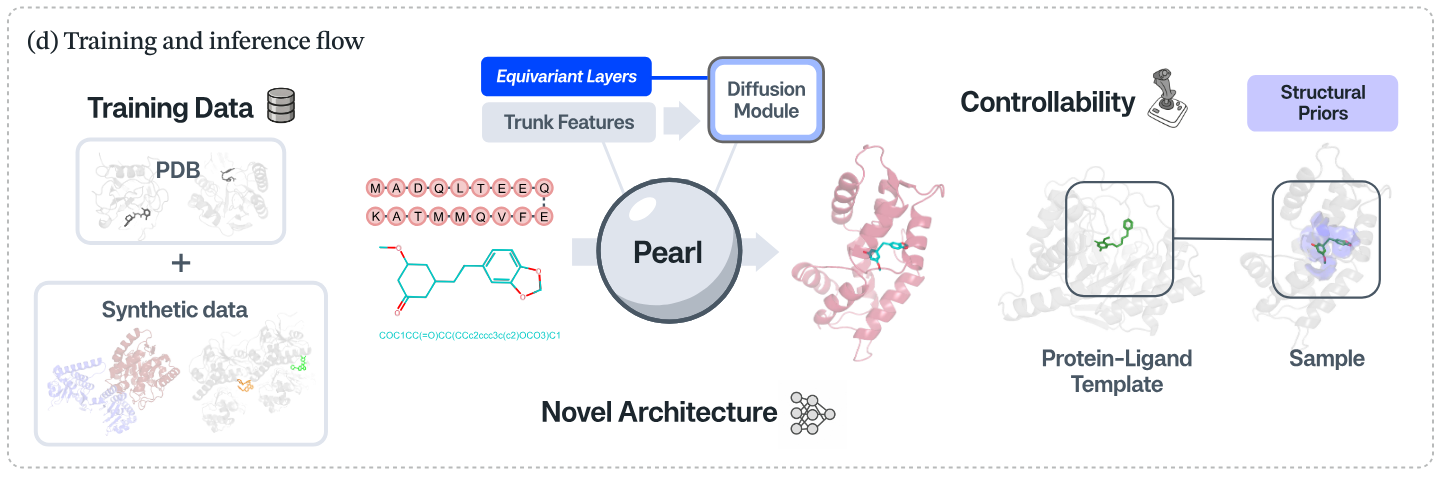
Pearl's training and inference flow, from technical report "Pearl: A Foundation Model for Placing Every Atom in the Right Location"
Pearl offers two modes that match real discovery workflows:
- In “cofolding” mode, it predicts a 3D binding pose from a protein sequence and a 2D ligand—useful early, before a pocket is well characterized.
- In “conditional” mode, scientists can steer predictions with prior knowledge such as a known pocket, cofactors, or related ligands; this in-context conditioning is designed to lift accuracy on flexible or otherwise difficult targets.
Pearl also slots into the company’s Genesis Exploration of Molecular Space (GEMS) platform alongside generative molecule design and property prediction. GEMS includes the following parts: language models conditioned on ADME to propose drug-like chemotypes diffusion models that generate 3D structures/poses; and physics-informed ML coupled to molecular dynamics and quantum chemistry for potency/selectivity and property prediction—all surfaced to chemists through the Nucleus interface for molecule generation, predictive screens, and review.
Topic: AI in Bio