AI Framework Produces Functional Nanobodies with Reduced Screening
A new preprint from the Brian Hie and Xiaojing Gao labs, in collaboration with the Arc Institute, introduces Germinal, a generative AI framework designed to accelerate antibody creation. Applied to nanobodies, it reduces the need for large-scale laboratory screening by combining AlphaFold-Multimer, which predicts protein–protein structures, with IgLM, a language model trained on antibody sequences. The system is reported to generate functional antibodies with far fewer experiments than conventional methods, potentially making custom antibody design more accessible for both research and therapeutic applications.
Obtaining antibodies that bind precisely to chosen targets has traditionally required either immunizing animals or screening thousands to millions of random variants. Computational methods have been explored as an alternative, but success rates have generally been too low to avoid large experimental campaigns.
Germinal approaches this differently by integrating both sequence and structure. AlphaFold-Multimer provides structural predictions, while IgLM biases designs toward sequences that resemble natural antibodies. The team added antibody-specific constraints so that binding occurs in the flexible complementarity-determining regions (CDRs) rather than rigid framework regions, and to prevent structural features unsuited for binding. This design strategy allows Germinal to produce antibodies that are both structurally precise and biologically realistic.
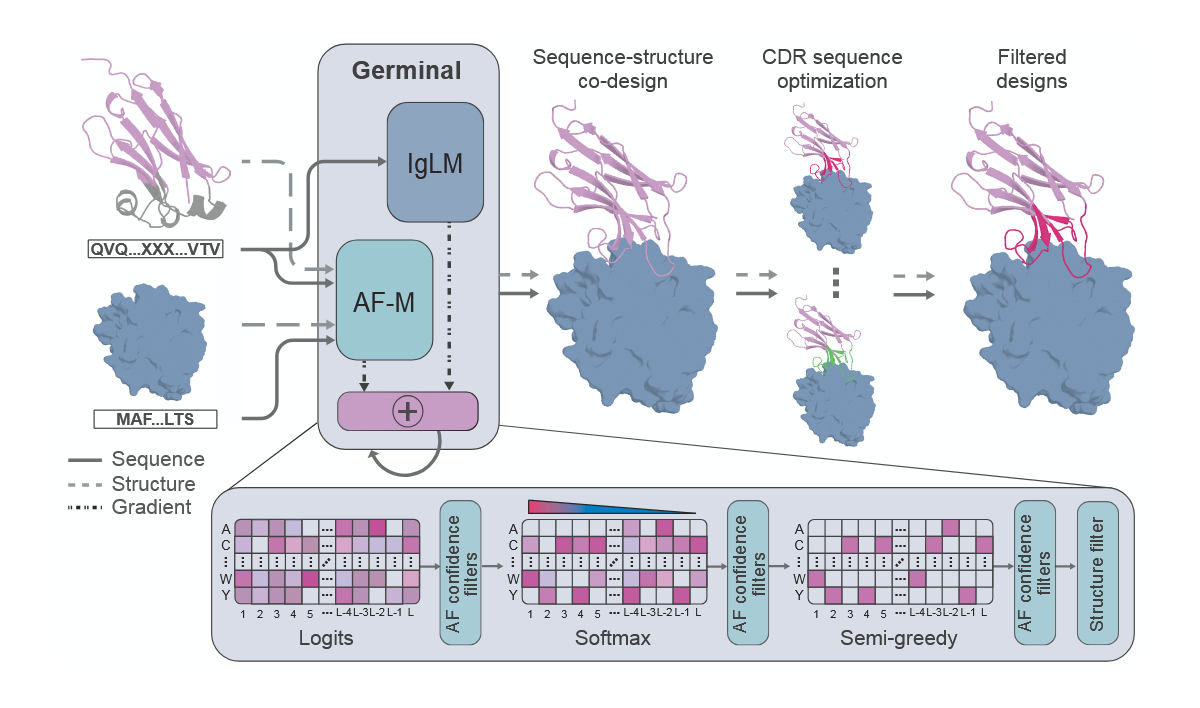
Efficient antibody generation via joint optimization of AlphaFold-Multimer and IgLM with Germinal, from preprint
In benchmarking experiments against four different protein targets, Germinal tested only 43–101 designs per antigen and achieved success rates ranging from 4% to 22%. The strongest binders reached nanomolar affinity, with one design binding at 140 nanomolar, a level approaching therapeutic relevance. Importantly, the nanobodies were shown to express robustly in mammalian cells.
The team validated designs through a two-step testing process: a rapid cell-based screen using split-luciferase assays, followed by detailed biophysical measurements on the strongest candidates. This pipeline allowed them to move from computational proposals to experimentally confirmed binders more efficiently than typical workflows.
The researchers have released Germinal’s code and protocols as open source, aiming to make the framework broadly available to both academic and industrial groups. The supplementary data can be accessed via the preprint.
We track developments like this weekly in Where Tech Meets Bio—our newsletter on startups, platforms, and deals at the intersection of biotech and digital.
Topic: AI in Bio