BenchSci Reports New System to Extract Experimental Context From Biomedical Papers Into a Traceable Evidence Graph
BenchSci reports a method to convert biomedical papers into structured, evidence-linked data intended to support a Biological Evidence Knowledge Graph and neuro-symbolic reasoning across disease biology. The system is described as operating across domains and at throughput measured in thousands of papers per day, with evaluations involving roughly fifty domain experts, according to the company.
BenchSci names the approach Literature Extraction and Network Semantics (LENS) and presents it as a two-phase pipeline.
- The first phase “deep reading” turns full text into machine-readable structure aligned to a stated universal scientific grammar—extracting research questions, methods, measured results, and conclusions with provenance.
- The second phase “knowledge synthesis” links those elements into inferential networks, normalizes entities and synonyms, preserves pathway connections, and maintains evidence chains from methods to conclusions. The company positions this as evidence-first extraction of experimental context rather than claim summarization.
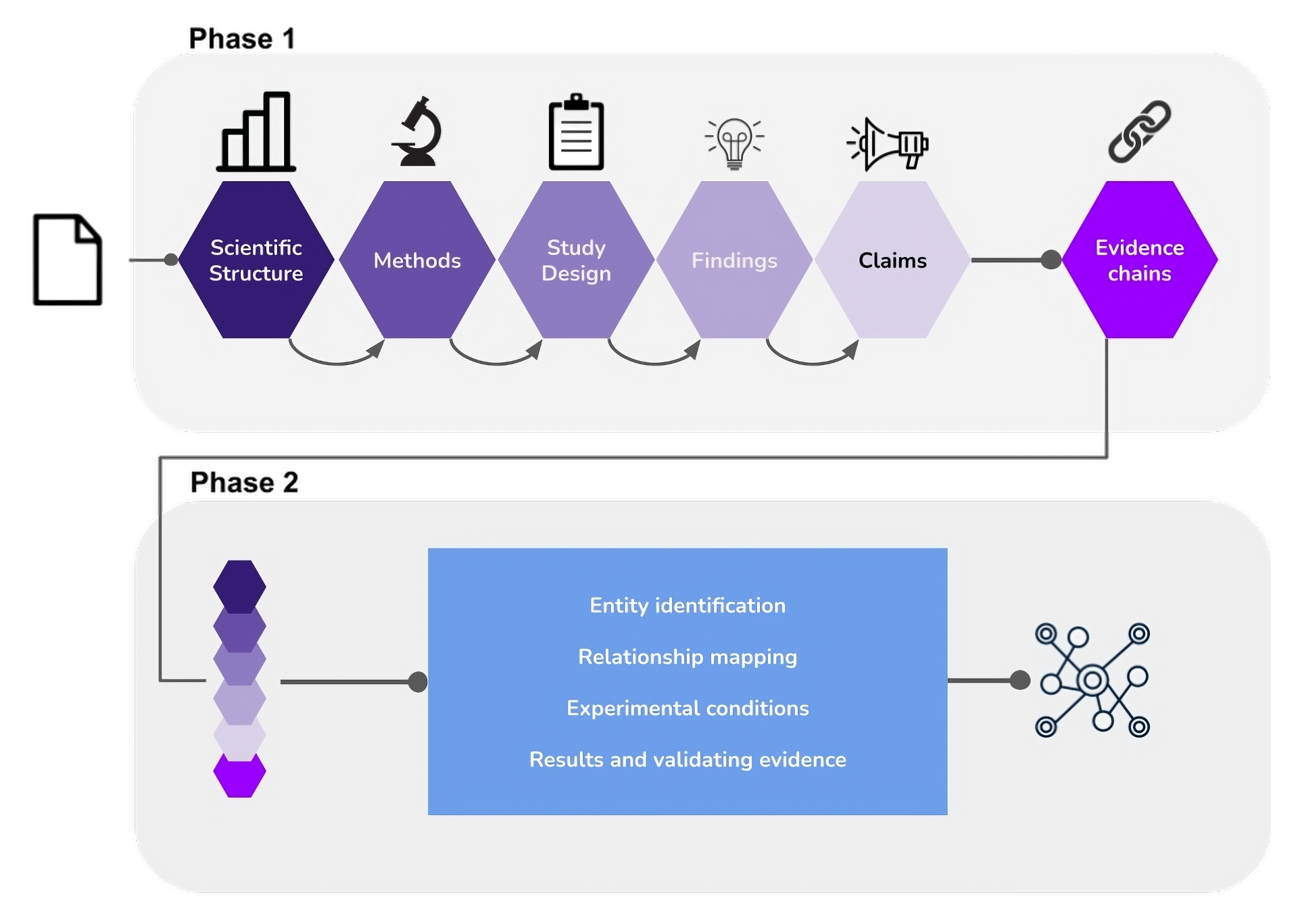
Phases 1 and 2; BenchSci, Fernando Suarez
Performance is reported as over 90% completeness with under 1% hallucination across biomedical domains. BenchSci contrasts this with general-purpose language models that, per the blog’s attribution to independent academic benchmarks, capture an estimated 15-35% of critical biomedical concepts in naive, zero-shot settings and, in the company’s internal conversational tests, achieve 32.5% completeness for GPT-4 and 35% for Claude. BenchSci also notes that domain-specific systems can reach 80-90% accuracy in narrow scopes and that manual curation operates at a pace of papers per week rather than thousands per day.
See also: From Data to Discovery: How BenchSci’s ASCEND Builds a Map for Biomedical Reasoning
The stated objective is to enable a Biological Evidence Knowledge Graph as a living, traceable map of disease biology spanning genomics, proteomics, metabolomics, preclinical and clinical evidence. BenchSci frames this as the substrate required for neuro-symbolic AI to infer causal relationships, generate evidence-based hypotheses, and support decisions with full source traceability. The company says the system works across new research areas without domain-specific retraining.
Topic: AI in Bio