Human Cells Edited at Megabase Scale With Bridge Recombinases
Researchers report in Science a genome-editing approach that rearranges long stretches of human DNA—insertions, deletions, and inversions—within regions up to about one megabase (with demonstrations of ~920 kb inversions and ~130 kb deletions), with reported insertion efficiencies up to 20% and 82% on-target specificity.
This work was initially published as a preprint on bioRxiv on May 16, 2025, and includes demonstrations such as excising repeats in constructs modeling Friedreich’s ataxia and deleting the BCL11A enhancer, the same target disrupted in an FDA-approved sickle cell therapy; plans include testing in immune and stem cells and engineering variants beyond one megabase.
Arc Institute’s team, led by Patrick Hsu with co-lead Silvana Konermann and first author Nicholas Perry, optimized a “bridge recombinase” system for human cells after screening 72 natural candidates from parasitic mobile genetic elements (“jumping genes”); about 25% showed some activity, but only ISCro4 warranted further engineering.
According to Arc’s press text, systematic protein and guide optimization yielded up to 20% insertion efficiency and 82% specificity at intended sites. Bridge recombinases pair a DNA-rearranging enzyme with a structured “bridge RNA” that binds two genomic sites, enabling one-step insert, excise, or flip operations. Commentary accompanying the release highlights how this approach does not rely on cellular DNA repair pathways and highlights reported inversions up to ~920,000 bp and deletions around ~130,000 bp with no apparent distance dependency.
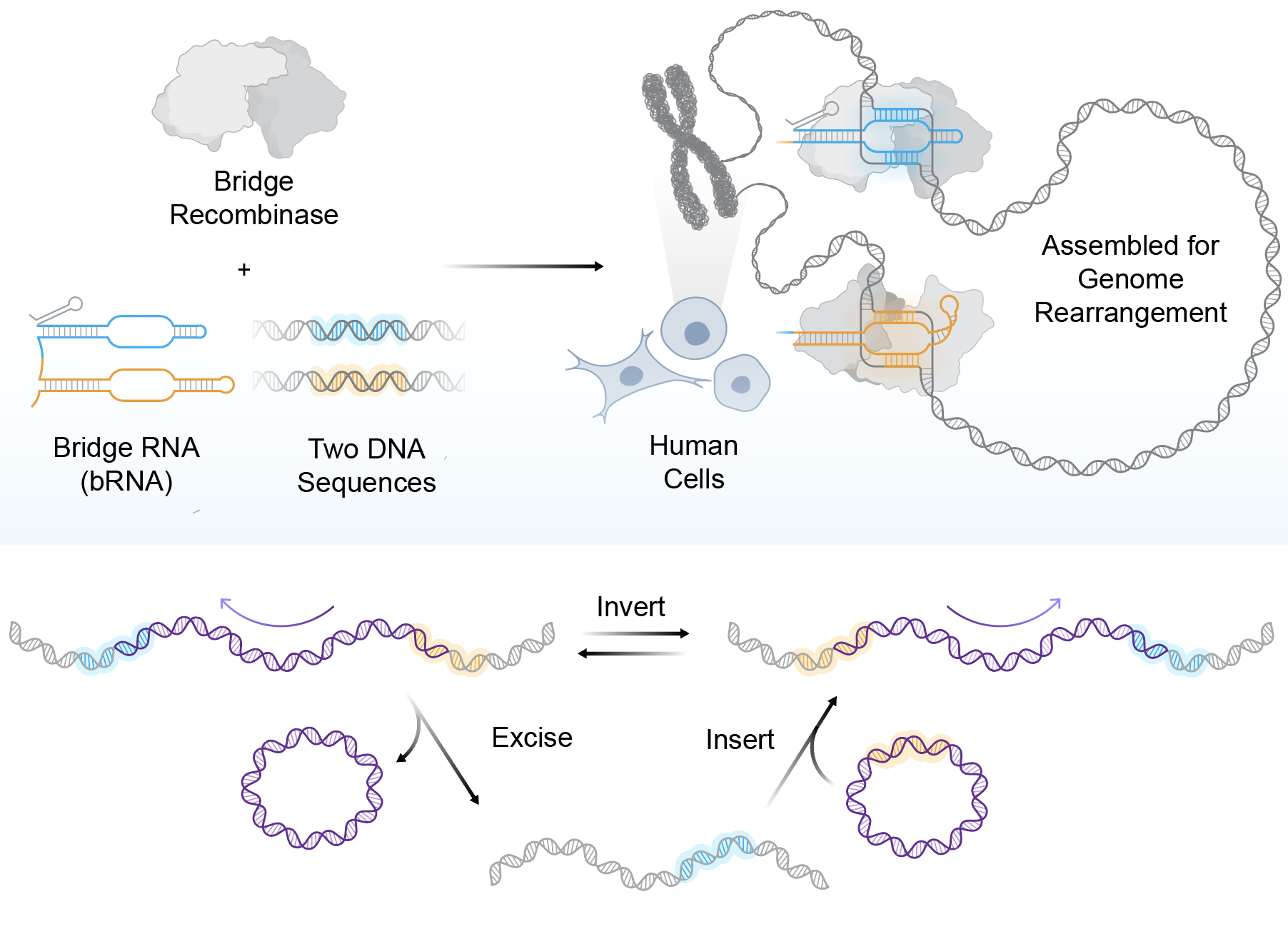
Bridge recombinases use dual-targeting to insert, delete, or invert DNA segments in a single programmable step. Credit: Chiara Ricci-Tam/Arc Institute.
In proof-of-concepts, the team removed expanded GAA repeats from artificial constructs modeling Friedreich’s ataxia (where healthy individuals carry fewer than 10 repeats versus up to ~1,700 in patients) eliminating over 80% of expanded repeats in some constructs, and deleted the BCL11A enhancer, the same target disrupted in an FDA-approved therapy. The group frames applications across repeat-expansion disorders (with Huntington’s cited as a potential class) and for modeling cancer-linked large chromosomal rearrangements. According to the press release, the system may be deliverable via RNA alone.
The study builds on prior work introducing bridge recombinases in bacteria and lists contributions from Arc/UC Berkeley (Hsu lab), Stanford (Konermann), and the University of Tokyo (Hiroshi Nishimasu) on structural and mechanistic aspects. Arc states that plasmids are available via Addgene along with a design tool that generates bridge RNA sequences from user-provided genomic pairs. While the platform is described as capable of rearrangements up to ~1 Mb, the demonstrated human-cell edits in this paper reached ~920 kb inversions and ~130 kb deletions; next-generation systems aiming beyond one megabase are described as in development.
Earlier this month, a Stanford–Arc team reported AI-designed bacteriophages: using their Evo/Evo2 genome models, they proposed about 300 variants of the small φX174 phage, and 16 worked in E. coli and killed bacteria. The result points to practical whole-genome design for small viruses with possible uses in phage therapy and delivery vectors, while also raising clear biosafety questions; the authors said their training data excluded human-infecting viruses.
Citation: Perry et al., “Megabase-scale human genome rearrangement with programmable bridge recombinases”, Science (2025), DOI: 10.1126/science.adz0276.
We track developments like this weekly in Where Tech Meets Bio—our newsletter on startups, platforms, and deals at the intersection of biotech and digital.
Topic: Next-Gen Tools