AI Model Built on Transformer Architecture Maps 1,300 Mouse Brain Regions in Data-Driven Atlas
Researchers at UCSF and the Allen Institute have developed a transformer-based AI model CellTransformer that maps 1,300 brain regions and subregions in the mouse brain, including previously uncharted subregions. The model was applied to large-scale spatial transcriptomics datasets, results were published October 7 in Nature Communications.
CellTransformer applies the transformer architecture common to language models like ChatGPT but adapts it to biological spatial data. While large language models learn from word sequences, this model learns from spatial relationships between cells. According to UCSF's Reza Abbasi-Asl, it leverages this context to build up an understanding of tissue structure at scale.
The system analyzes how cells are arranged in physical space and predicts their molecular features by learning from their local cellular neighborhoods. This approach enables the model to generate a fine-grained map of brain architecture based entirely on data, without relying on expert-drawn anatomical boundaries. The resulting parcellation reportedly matches known regions like the hippocampus and reveals new subdivisions within under-characterized areas, such as the midbrain reticular nucleus, which plays a complex role in movement initiation and release.
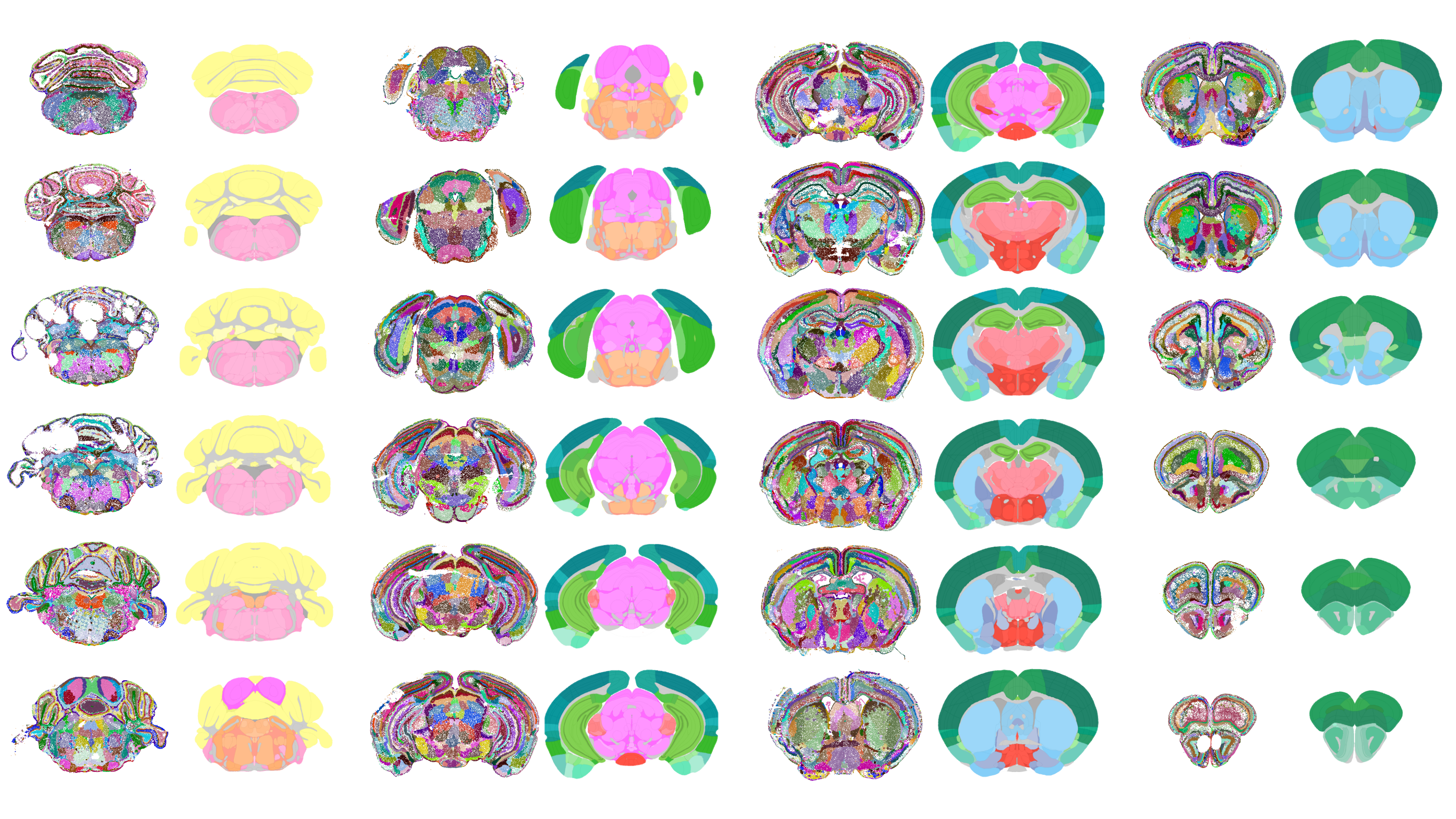
Examples from 1300 regions/subregion in mouse brain created by CellTransformer(Credit: University of California, San Francisco)
Validation against the Allen Institute’s "gold standard" Common Coordinate Framework (CCF) showed a high degree of alignment between model-defined regions and expert-defined anatomical areas. First author Alex Lee noted that this agreement strengthens the likelihood that newly predicted subregions are biologically relevant, pending further validation.
See also: Neuroscientists Map Cubic Millimeter of Mouse Brain
Unlike previous brain atlases focused on cell types or human-defined boundaries, this map defines anatomical subregions based on cellular and molecular data. The authors describe this as one of the most granular and complex data-driven brain maps for any animal to date.
The developers also suggest the method is tissue-agnostic. It could potentially be applied to other organs or diseases like cancer, wherever spatial transcriptomic datasets are available.
Topic: AI in Bio